Application Settings
The following sections are written for Persephone administrators.
The application settings for Persephone are stored in two places: the Persephone.exe.config file deployed with Persephone and a file config.jsv that resides in the ClickOnce application's root directory. The user settings, that the users can change based on their preference, must be specified in Persephone.exe.config:
<userSettings>
<Persephone.Properties.Settings>
<setting name="AutoArrangeAddingPlate" serializeAs="String">
<value>True</value>
</setting>
<setting name="AutoArrangeClosingPlate" serializeAs="String">
<value>True</value>
</setting>
<setting name="WheelZoomIncrement" serializeAs="String">
<value>5</value>
</setting>
...
When the ClickOnce application is launched for the first time, the parameters get their default values from the config file until the user decides to change them via Tools/Settings form. Please note, that after that, the modified values will persist on the user's disk, and further changes to the config file will have no effect.
The other parameters, such as the server connection strings or visibility of certain components of the application, that are common for all users, should be specified in config.jsv file. Place this file into the same directory as Persephone.application, the starter file given to the users via the ClickOnce URL, such as, for example, http://yourcompany.com/persephone/Persephone.application.
The file Persephone.exe.config comes pre-configured and, hopefully, does not require any modification before the deployment. As long as it is a part of the ClickOnce deployment, it is digitally signed. This means that if you change anything in its text, you must re-sign the package using mage utility. To avoid these complications with other settings, that
The settings are first read from the file config.jsv. If the required settings are missing in that file, they will be read from Persephone.exe.config. The newer approach highly recommends using the file config.jsv, as it can be changed by the admin person and will have an effect
The Persephone.exe.config file is an XML file organized in section groups and sections containing user-configurable settings. The following are section groups and sections that you may want to update for your deployment.
Some application configuration is stored in the ORGANISM_CONFIG database table. For example, it enables you to define the hyper-links for various gene qualifiers or specify which qualifier contains the gene name or function.
ORGANISM_CONFIG Table — Qualifier Links
A gene may contain annotation in a form of "qualifiers" that are stored as key-value pairs in the GENBANK_ANNOT_QUALIFER table. This table has three columns: ANNOT_ID, QUALIFIER, and VALUE. The QUALIFER column serves as the key and the VALUE column contains the qualifier value. The key-value pairs are displayed in the Annotation Details dialog:
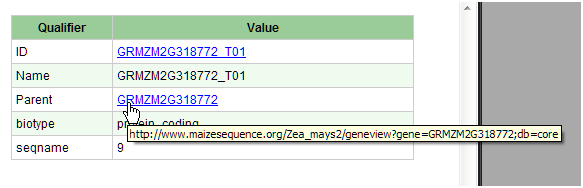
To convert the value to a URL, add records to ORGANISM_CONFIG table with APP_FEATURE set to "QUALIFIER_LINK".
For example, a gene may have a QUALIFIER='ID' and VALUE='GRMZM2G130095_T03' recorded in the GENBANK_ANNOT_QUALIFIER table. Add a record to ORGANISM_CONFIG where APP_FEATURE='QUALIFIER_LINK', CONFIG_KEY='ID' and CONFIG_VALUE='http://www.maizesequence.org/Zea_mays2/transview?transcript=%s'.
Please note the “%s” substring will be replaced in the program with the actual value of the qualifier to complete the URL that will become:
http://www.maizesequence.org/Zea_mays2/transview?transcript=GRMZM2G130095_T03
When shown in the grid, a qualifier can be renamed. The hyperlink can also have a custom label:
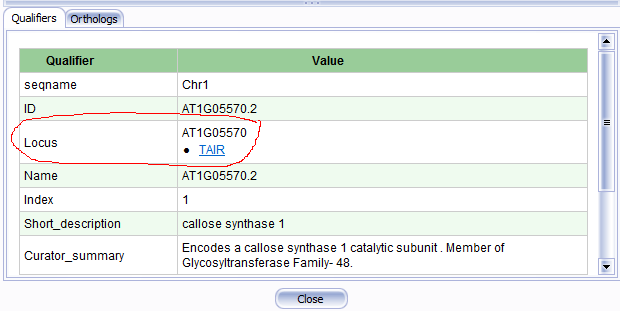
For example, to replace a qualifier "Parent" with the text "Locus" and give the link a custom label "TAIR", insert a record into ORGANISM_CONFIG that for CONFIG_KEY="Parent" has QUALIFIER_LABEL="Locus" and LINK_LABEL="TAIR".
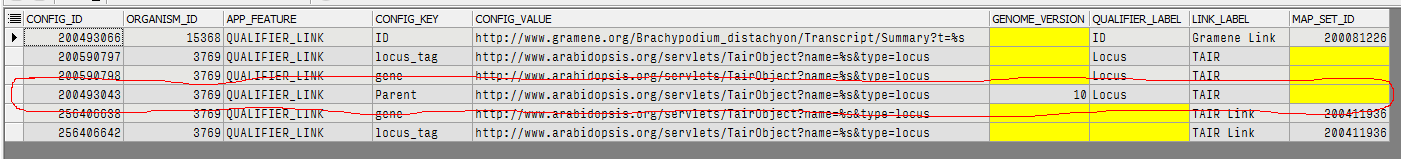
ORGANISM_CONFIG Table — Annotation Search
The Annotation tab in the Search dialog includes an option to search annotations by Gene Name, Gene Function.
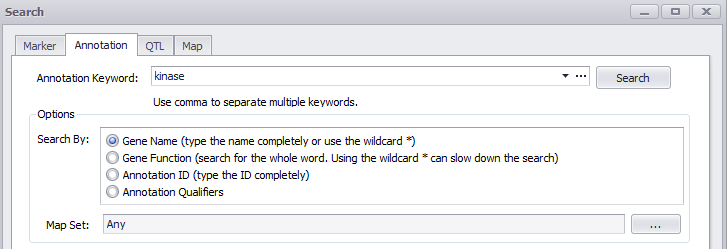
Search for Gene Function
The table ORGANISM_CONFIG should contain records that specify which qualifiers in a given organism contain gene name or gene function. For example, Arabidopsis annotation has the gene name in 'locus_tag' qualifiers. This is recorded as an entry in ORGANISM_CONFIG, where
APP_FEATURE='ANNOTATION_SEARCH'
CONFIG_KEY='GeneName'
CONFIG_VALUE='locus_tag'
Sometimes the gene names (or gene functions) are stored in several different qualifiers. To enable the search through all such qualifiers you need to list them all in ORGANISM_CONFIG. For the function search the CONFIG_KEY should be set to 'GeneFunction'.
ConnectionString Section
The encrypted database connection string is stored in the value attribute of this section. Use the cipher program, which is also used in Persephone, to encrypt a connection string and use it as the value attribute of this section.
<ConnectionString value="-encrypted by cipher.exe-" />
WebBrowserTab Section
The first tab page in the main window of Persephone contains a Web Browser control that can be configured to display any Web content. Settings that apply to this tab page can be found under the WebBrowserTab section. The WebBrowserTab section contains key-value pair information that relates to this tab page. There are three key-value pair information that is used by Persephone to determine how the tab page is displayed: TabName, URL, and IsSelected. Here is an example of how to use the WebBrowserTab section.
<WebBrowserTab>
<add key="TabName" value="Company News" />
<add key="URL" value=http://www.company.com/News/News-2018-Releases.html />
<add key ="IsSelected" value="false"/>
</WebBrowserTab>
The following table contains key and click combinations:
|
Key |
Value |
|
TabName |
Specifies the label to use for the tab page. |
|
URL |
Specifies the URL of the Web content the WebBrowser control will load. |
|
IsSelected |
Set to true to make the tab page the default active page or false to make the Map Alignment page the default active page. |
BlastSettings Section Group
This section group contains setting for configuring the real-time BLAST feature of Persephone. The typical BlastSettings section is as follows:
<BlastSettings>
<!-- If true, use NCBI instead of WU-blast. -->
<UseNcbiBlast value="true" />
<UseNewBlastForm value="true" />
<!-- New-style BLAST configuration; talks to a dedicated Cerberus instance -->
<NcbiBlast
WebApiUrl="http://company.com/cerberus"
MultipleSubjects="true"
LengthWarning="0">
<BlastP>
<add key="Fast" value="-seg "yes" -evalue 1e-6 -word_size 4 -threshold 999 -culling_limit 10" />
<add key="Normal" value="-seg "yes" -evalue 1e-6 -threshold 999 -culling_limit 10" />
</BlastP>
<BlastX>
<add key="Fast" value="-seg "yes" -evalue 1e-6 -word_size 4 -threshold 999 -culling_limit 10" />
<add key="Normal" value="-seg "yes" -evalue 1e-6 -threshold 999 -culling_limit 10" />
</BlastX>
<BlastN>
<add key="Fast" value="-dust "yes" -evalue 1e-10 -word_size 28 -culling_limit 10" />
<add key="Normal" value="-dust "yes" -evalue 1e-10 -word_size 20 -culling_limit 10" />
<add key="Primer" value="-evalue 1000 -word_size 20" />
</BlastN>
<TBlastN>
<add key="Fast" value="-seg "yes" -evalue 1e-6 -word_size 4 -threshold 999 -culling_limit 10" />
<add key="Normal" value="-seg "yes" -evalue 1e-6 -threshold 999 -culling_limit 10" />
</TBlastN>
</NcbiBlast>
</BlastSettings>
- The WebApiUrl field should point to a Cerberus instance, as the BLAST search is now mediated by Cerberus. It passes the query to its local copy of BLAST and returns the results back to Persephone.
Update:
The following section is obsolete. See BLAST section in Cerberus documentation.
The Red Hat Linux webpost program (wrapped into a script webpostn.s to set environment variables) accepts HTTP POST request from Persephone, runs BLAST, and returns the response back to Persephone. The text of this script is shown below:
#!/bin/tcsh
#specify home location for ncbi blast
set BLAST_HOME=/ceres/process/v_0.0/bin/Linux/ncbi_blast
#When set, this file will contain the command line parameters with timestamps.
setenv WEBPOST_LOG /tmp/blastlog2
setenv WEBPOST_TEMPDIR /mounted/tmp
setenv PATH $BLAST_HOME/bin\:$PATH
setenv BLASTMAT $BLAST_HOME/data
/webpostn
- $WEBPOST_LOG – Log file that will keep all the command line parameters and timestamps of the BLAST jobs.
- $WEBPOST_TEMPDIR – Alternative location of temporary sequence files (default: /tmp).
Both webpostn and webpostn.s should be placed in web server's cgi-bin directory (or other executable directory). If you go to URL of webpostn using a browser, the page should look something like this:
Error: No sequence
