BLAST Results Alignment
If you ran BLASTN or TBLASTN, the Alignment tab will be available as shown below. (The results for BLASTN are shown.)
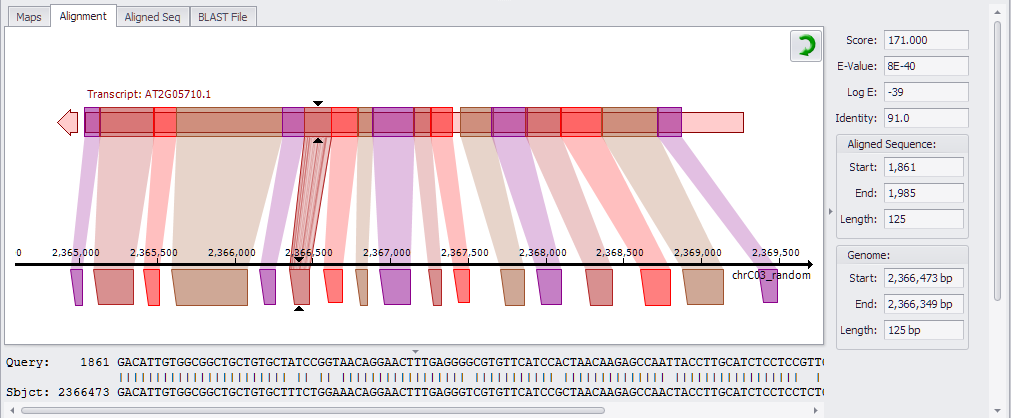
The Alignment tab lets you visualize gaps in the alignment. By default, the Alignment tab shows a horizontal view with the genomic sequence shown on the bottom and the Aligned Sequence with adjusted orientation is displayed on the top. The width and color intensity depend on the score of the match.
If you highlight a different row in the HSPs panel above, this will update the chart to the corresponding data. Click the arrow to toggle between horizontal and vertical views (see the figure below).
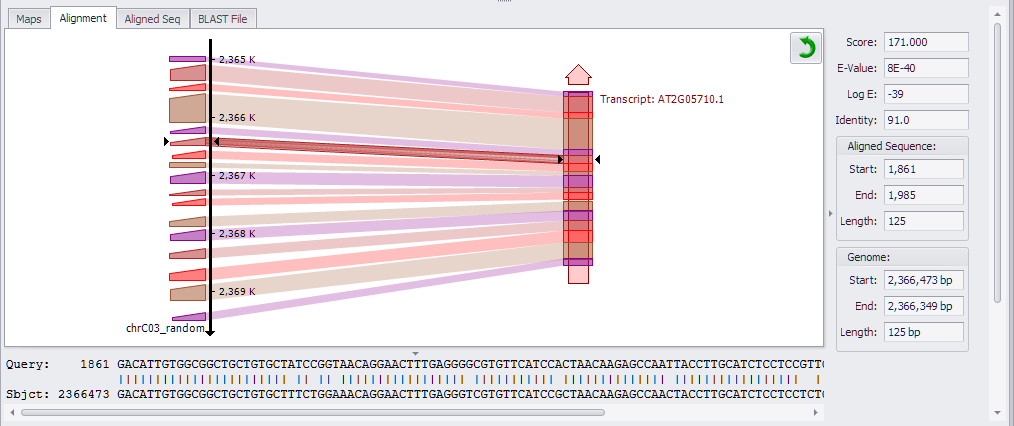
Note
It is sometimes important to see which part of the query is aligned to the chromosome
is aligned to the chromosome . For example, in the case of duplicated genes
. For example, in the case of duplicated genes , the same portion of the protein query can match several regions of the genome.
, the same portion of the protein query can match several regions of the genome.
