BLASTX
Follow the steps below to run the BLASTX program, which performs a nucleotide (translated in all six frames by the program) versus protein comparison.
- View the details form for some gene:
- Click the Spliced Sequence tab.
- Click Run BLAST... button. The Run BLAST window displays.
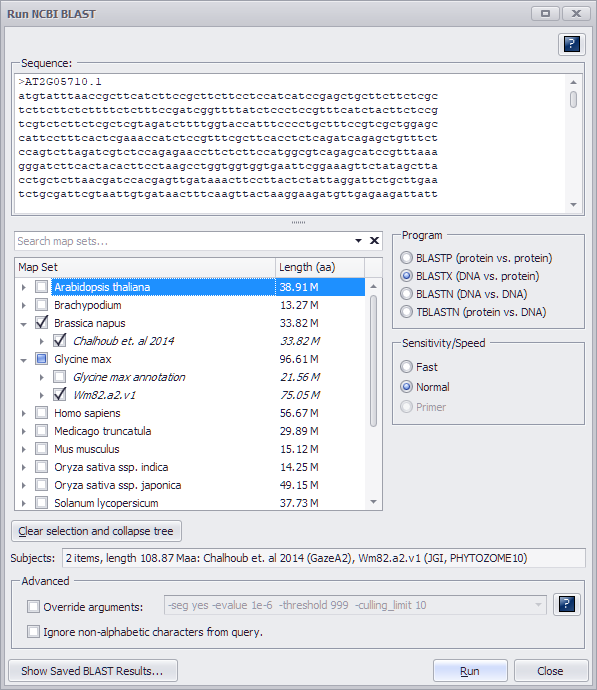
- The Sequence text box will be filled with the spliced sequence of the selected gene model
Note
The FASTA header line is optional if only one sequence is used as a query. In case of multiple query sequences each of them should have the FASTA header. The identifier from the header will be shown in the Query column of the blast results window, so please keep the identifiers unique.
header line is optional if only one sequence is used as a query. In case of multiple query sequences each of them should have the FASTA header. The identifier from the header will be shown in the Query column of the blast results window, so please keep the identifiers unique.
- In the Program panel check (select) the BLASTX radio button.
- Select the genome(s) BLAST file to use from the map set tree panel.
- Check (select) the Fast or Normal radio button in the Sensitivity/Speed panel. (In the figure above Normal was selected.)
Note
The Primer option is grayed out since it is not a valid option for BLASTX.
- Optional. Check (select) the Override arguments in Advanced panel if the default parameters need modification. Please note, there is a "?" button that opens the help page with the list and description of BLAST parameters.
- Click OK to run. The BLAST in Progress window displays.
- When the BLAST process finishes, the BLAST Results window displays. See BLAST Results Window for details.
