Adding Samples
Sample information for genotyping studies can be loaded from either VCF or delimited text (e.g., CSV) format files. A delimited text file format should contain each sample in its own row as shown below.
|
Source1 |
DNA_UNIQUE_ID |
DNA_VARNAME_source |
DNA_Othername_source |
ORI_COUNTRY |
Genetic_Stock_Accno |
Variety Group (Tree) |
|
MC |
B001 |
黑标 |
Heibiao |
China |
I1A12996 |
Temperate japonica |
|
MC |
B002 |
三穗锦 |
Sansuijin |
China |
I1A28066 |
Temperate japonica |
|
MC |
B003 |
早生白 |
Zaoshengbai |
China |
I1A28115 |
Temperate japonica |
The genotyping sample names usually correspond to the samples listed in VCF files on the #CHROM line. Typically, you first load the variants from a VCF file ('add variants') and then add the extra information about the samples ('add sample') - make sure that SampleNameAddMode is set to AddOrUpdate, so that the new information is appended to the samples already existing in the database.
It is also possible to create the sample records first and then add the data from VCF files.
The general form of the command is
add sample -c <control.ini> [-t|-v]
As with the majority of PersephoneShell commands, first, test the instructions using the switch -t and then load the data with -v flag.
Text files
Here is an example of the control file for loading the extra sample information provided in a CSV file (please read the comments):
[ProcessRun]
; Run description: if specified, a custom description will be used. Will be ignored if a RunId is specified.
; otherwise, "Added sample information for {MapSet Accession No.}." will be used.
RunDescription="Added sample information for 3K RG from https://s3-ap-southeast-1.amazonaws.com/oryzasnp-atcg-irri-org/3kRG-phenotypes/3kRG_PhenotypeData_v20170411.xlsx(CAAS tab)"
[MapSet]
; Either MapSetId or MapSetPath is required.
; MapSetId: id of a target map set.
;MapSetId=232287170
; MapSetPath: path of a target map set.
MapSetPath=/Rice-platinum standard/Os GJ-temp: IRGSP-1.0 (Nipponbare)
[Sample]
; Source (required): a TXT file located locally or remotely accessible via URL.
Source="$DATA/rice/3KsampleInfoCAAS.csv"
; Number format culture: specifies a culture name used to parse numbers in data. Default value is en - English.
; e.g. de - German, es - Spanish, fr - French. For more cultures, https://msdn.microsoft.com/en-us/goglobal/bb896001.aspx
;NumberFormatCulture="fr"
; AddModes: choose a mode to add name or qualifiers among
; 1. AddAnyway: Add regardless of duplication. Faster as it does not check.
; 2. AddOrDie: add if not exists; die (throw exception) otherwise.
; 3. AddOrUpdate: add if not exists; update otherwise.
; 4. AddOrSkip: add if not exists; skip otherwise.
SampleNameAddMode=AddOrUpdate
SampleQualifierAddMode=AddOrUpdate
; FileType: {Text (delimited text file), Vcf}
FileType=Text
;---------------------------------------------------------------------------------------------------------------
; Parsing Information
;---------------------------------------------------------------------------------------------------------------
; SkipHeaderLines: the number of lines to skip parsing
TextSkipHeaderLines=1
; CommentPrefix: comment prefix to skip parsing
;TextCommentPrefix="#"
; Delimiter: specify one among Colon(:), Comma(,), Period(.), Hyphen(-), SemiColon(;), Slash(/), Tab(\t), VerticalBar(|)
TextDelimiter=Comma
;-------------------------
; SampleNameIndex (required): column index(0-based) for sample names.
TextSampleNameIndex=1
; SampleDescriptionIndex: column index(0-based) for sample description.
;TextSampleDescriptionIndex=3
; SampleQualifierIndex: column index(0-based) for additional information.
; TextSampleQualifierIndex.INDEX(0-based)=qualifierName((:displayText),dataType,dataFormat)
;TextSampleQualifierIndex.3="Description"
TextSampleQualifierIndex.0="Source"
;TextSampleQualifierIndex.4="DNA_Accno_source"
TextSampleQualifierIndex.2="DNA_VARNAME_source"
TextSampleQualifierIndex.3="DNA_Othername_source"
TextSampleQualifierIndex.4="ORI_COUNTRY"
TextSampleQualifierIndex.5="Genetic_Stock_Accno"
;TextSampleQualifierIndex.13="depth","Double"
TextSampleQualifierIndex.6="Variety Group (tree)"
VCF files
The sample information can also be parsed out of the VCF files.
The VCF sample shown below contains ##SAMPLE parameter lines in the header that correspond to sample names listed in the #CHROM line.
Note
The ID and Description parameters are defined in the VCF 4.2 specification.
##fileformat=VCFv4.2
##INFO=<ID=AN,Number=1,Type=Integer,Description="Total Allele Count">
##FORMAT=<ID=GT,Number=1,Type=String,Description="Genotype">
##SAMPLE=<ID=HG00096,Gender=Male,Description="Donor, parents and grandparents were all born in the United Kingdom">
##SAMPLE=<ID=HG00097,Gender=Female,Description="Donor, parents and grandparents were all born in the United Kingdom">
##SAMPLE=<ID=HG00099,Gender=Female,Description="Donor, parents and grandparents were all born in the United Kingdom">
##SAMPLE=<ID=HG00100,Gender=Female,Description="Donor, parents and grandparents were all born in the United Kingdom">
##reference=GRCh37
#CHROM POS ID REF ALT QUAL FILTER INFO FORMAT HG00096 HG00097 HG00099 HG00100
Specify qualifiers other than ID or Description in the VcfSampleQualifierKey parameter as shown below.
[Sample]
Source="Samples/Sample/1000genomes.vcf"
;NumberFormatCulture="fr"
FileType=Vcf
;##SAMPLE=<ID=HG00096,Gender=Male,Description="Donor, parents and grandparents were all born in the United Kingdom">
VcfSampleNameKey="ID"
VcfSampleDescriptionKey="Description"
VcfSampleQualifierKey.Gender="gender"
The record VcfSampleQualifierKey.Gender="gender" specifies which entry on the ##SAMPLE line ('Gender=Female') should be parsed out, and which qualifier should be created in the database that will store this information (lowercase 'gender').
The sample qualifiers are shown in Persephone's "Select samples" form. The user can select qualifiers from the list on the right and add columns to the grid, like shown below:
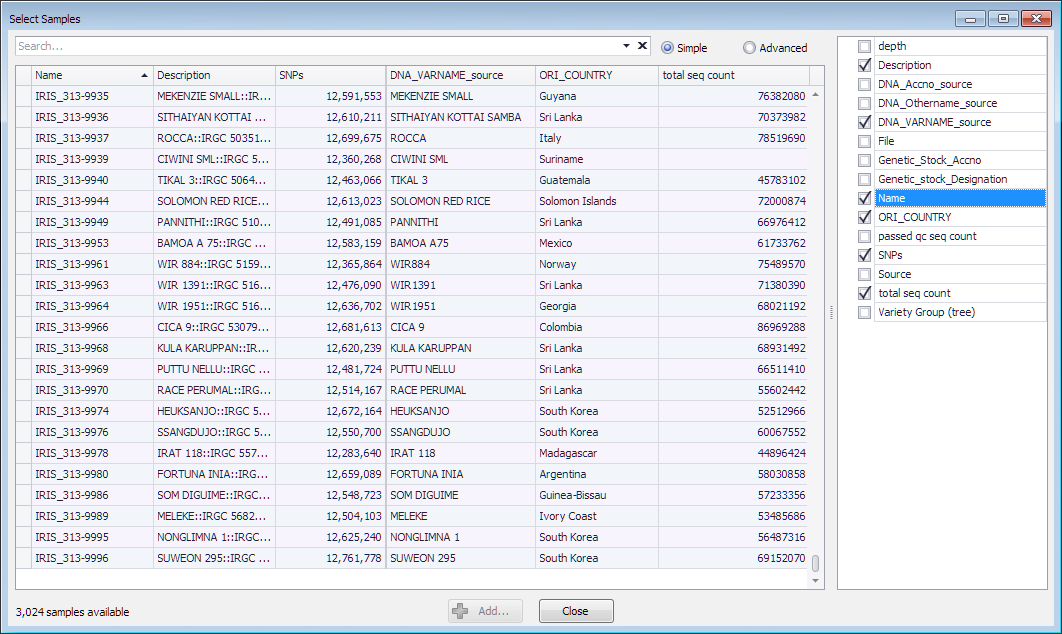
The qualifiers normally store strings, but other data types are also supported ('int','double'). For example, to store an integer qualifier 'total seq count' that is read from the record 'tsc' use this construct:
VcfSampleQualifierKey.tsc="total seq count",int
