Markers
A genetic marker is a genomic feature that typically occupies a defined position on one or more genetic or physical maps. A collection of such markers can be used to identify individuals, populations, or species, and may also assist in tracking inheritance patterns or mapping traits.
Genetic markers are foundational tools in fields such as genotyping, evolutionary biology, and molecular breeding — serving as reference points for comparative studies and genomic analysis.
Markers are often associated with specific DNA sequences, which can range from short flanking regions around single nucleotide polymorphisms (SNPs) to longer repeat elements like microsatellites. The marker object in the database is identified not only by its name but also by its source organism. Each marker may have multiple aliases, associated sequences, and functional qualifiers or annotations.
The figure below shows the hierarchy of markers in Persephone.
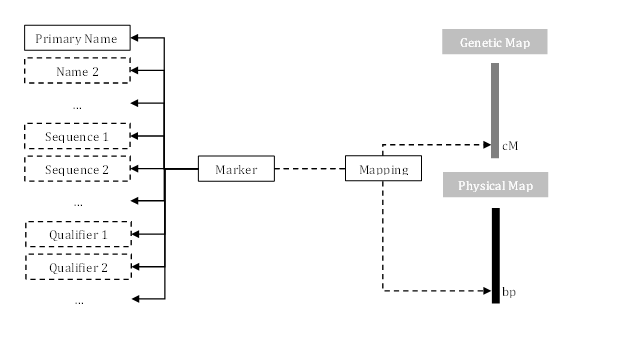
The following sections describe common control file sections used to add and modify markers.
Tip
See Add Markers for an example of using a control file to add markers.
MarkerNameType
A single marker can have multiple names. Each name has its own type. For example, a marker named 'adh2' could also be called as 'S45023' or 'X02915' elsewhere. If you want to consider adh2 as 'MARKER_NAME' and other names as 'ALIAS', you define those name types in the section [MarkerNameType] of the INI control file as shown in the example below. If name types MARKER_NAME and ALIAS already exist in the database, their description will be updated; otherwise, the types will be added.
The following shows the format for the [MarkerNameType] control file section.
[MarkerNameType]
MARKER_NAME="Primary marker name."
ALIAS="Another name used to call a marker."
In each marker track section, specify types of names and a primary marker name type given file source.
For example, if the adh2 marker is defined in a source GFF file as shown below:
chr1 . SNP 440159 440159 91.68 - . ID=adh2;Name=S45023;Alias=X02915
then corresponding track section of the control file would be
[MyMarkerTrack1]
PrimaryMarkerNameType=MARKER_NAME
GffMarkerNameAttributeKey.ID=MARKER_NAME
GffMarkerNameAttributeKey.Name=ALIAS
GffMarkerNameAttributeKey.Alias=ALIAS
This set of instructions tells that a marker will be created with a primary name MARKER_NAME copied from a GFF attribute ID. There will be other names for this marker of type ALIAS copied from attributes Name and Alias.
TextMarkerName
In case the data is provided in the form of text file, the instructions start with the word Text. If the marker names are defined as follows:
Map_name pos marker_name alias1 alias2
chr1 440159 adh2 S45023 X02915
then the entries in the control file would be:
TextMarkerNameIndex.2=MARKER_NAME
TextMarkerNameIndex.3=ALIAS
The number 2 here is instructing to read MARKER_NAME from the third column with the 0-based index 2.
MarkerSequenceType
The following shows the format for the control file's MarkerSequenceType section, which introduces the marker sequence types.
[MarkerSequenceType]
ASSAY_SEQ="The probe assay sequence of the marker."
The corresponding parsing instructions would look similar to the ones in the previous section:
; TextMarkerSequenceIndex: column index(0-based) for a marker sequence.
; The following means that the column 1 contains sequence of type "ASSAY_SEQ"
TextMarkerSequenceIndex.1=ASSAY_SEQ
AddMode
Whenever possible, try to reuse the markers that already exist in the database. This will allow linking their mapped positions on different maps with connector lines, which are always drawn between identical markers.
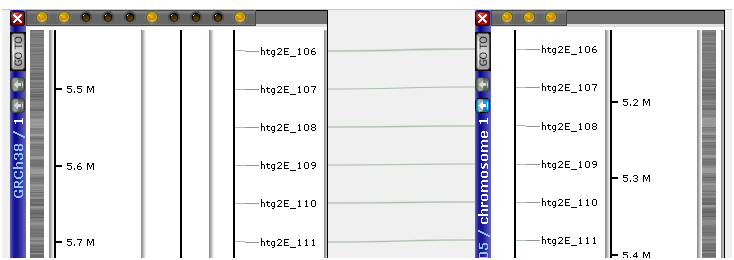
As we mentioned before, the identity of the marker is defined by the combination of its name and the source organism. So, if you see two markers with identical names that are not linked with a line, most likely, they have different SourceOrganismIds and were loaded as independent objects. If you want to reuse a marker, make sure when loading that it is assigned the same SourceOrganismId and use AddMode AddOrSkip.
|
AddMode |
Description |
|
AddAnyway |
Add regardless of duplication. Faster as it does not check. |
|
AddOrDie |
Add if does not exist; die (throw exception) otherwise. |
|
AddOrUpdate |
Add if does not exist; update otherwise. |
|
AddOrSkip |
Add if does not exist; skip otherwise. |
This modes are applicable to five different instructions during loading. Typically, MarkerNameAddMode is set to AddOrSkip to reuse the marker and MappingAddMode is set to AddAnyway to save time on checking on the existing mapping.
MarkerNameAddMode=AddOrSkip
MarkerSequenceAddMode=AddOrSkip
MarkerQualifierAddMode=AddAnyway
MappingAddMode=AddAnyway
MappingQualifierAddMode=AddAnyway
