Create
PersephoneShell can be used not only for loading the pre-processed data, but for generating new information, such as the orthologous relations between proteins/genes or mapping sequence tags.
The connectors between two aligned maps can be drawn based on ortholog pairs or common markers. PersephoneShell offers a command that will cut out small sequence tags from a reference genome, thus creating "fake" markers. They can be mapped onto another genome. This picture shows maps aligned by linking orthologous genes (left) or common markers (right) for two Pyrus species.
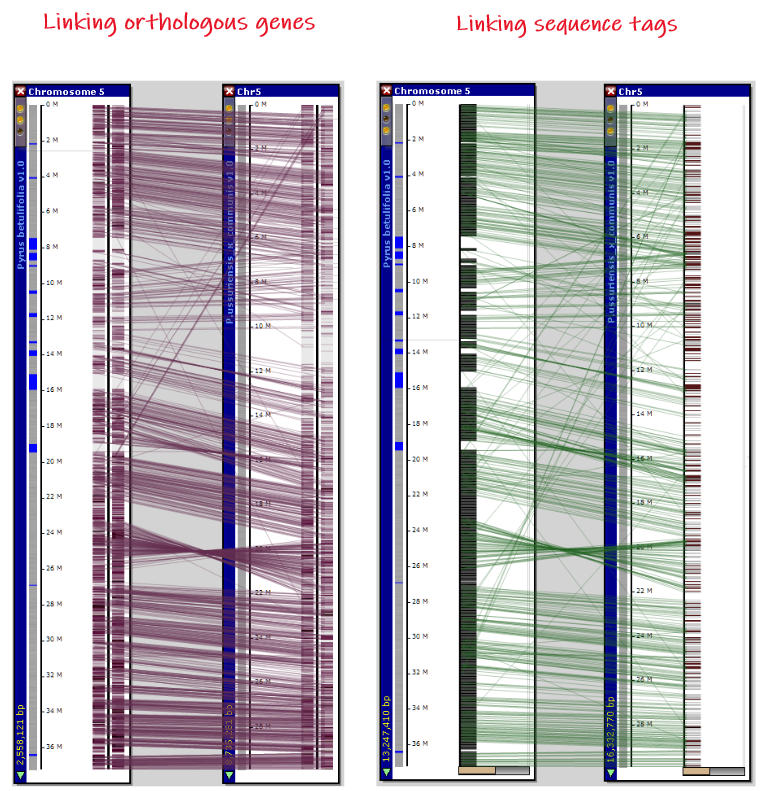
Using the tags can be beneficial if you compare genomes that show good sequence similarity. If this is the case, there is a good chance that the sequence tags from one genome will find a match on the other one. In the pan-genome scenario, where one deals with dozens of similar genomes, the process of mapping the tags can be much faster than finding the orthologs for each pair of genomes. When comparing the species with poor DNA similarity, it might be that using orthologous gene pairs discovered by a more sensitive protein sequence search could be the only option.
Create Function
