Edit translation code
By default, all DNA sequence translation into protein is using the standard translation code 1. For some organisms or organelles, the translation should be done with a different table. This can be assigned to individual maps or to all maps of a map set using the command
edit translation_code <mapset> [<code> [-f]]
The parameter <mapset> is given as MapSetId or path, as usual. Without other arguments, the command will list all the maps of the given map set with their corresponding translation code values. The code can be changed for individual maps or a range of lines (1..5):
PS> edit translation_code "Arabidopsis thaliana/TAIR10"
Editing map translation code of Arabidopsis thaliana/TAIR10
[##] MAP_NAME TRANSLATION_CODE
----------------------------------
[0] 1 1
[1] 2 1
[2] 3 1
[3] 4 1
[4] 5 1
[5] Mt 1
[6] Pt 1
Select maps by [lineNo] (0,2,10..20)? 6
Enter new translation code? 11
[##] MAP_NAME TRANSLATION_CODE
----------------------------------
[0] 1 1
[1] 2 1
[2] 3 1
[3] 4 1
[4] 5 1
[5] Mt 1
[6] Pt 11
Do you want to select other maps? (Y/N) N
Update the map translation codes in the database? (Y/N) Y
Map set translation codes updated
To update the translation code for all maps of the map set from the command line, add the translation code to the parameter list, optionally supplementing it with the force flag -f to skip the confirmation questions.
PS> edit translation_code "Arabidopsis thaliana/TAIR10" 1 -f
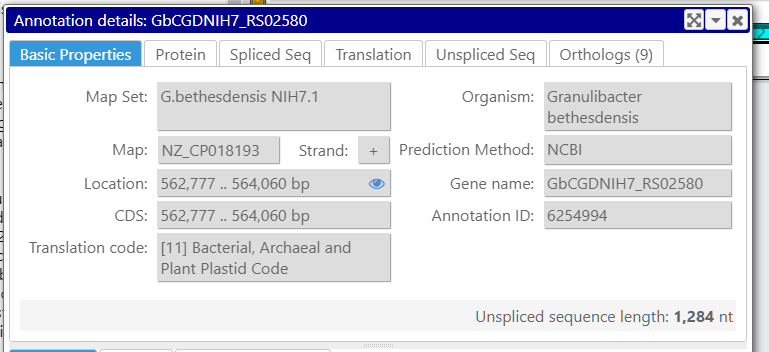
In Persephone, the translation code used for producing the protein sequence is listed in the gene properties form: