Edit chromosomes
As was noted in 'add sequences' section, some of the maps can also be designated as chromosomes. This helps correctly sorting the maps in the data grids, or deciding which maps should be shown as representatives of the full genome in the graphical output of various search results.
The logic in listing all the maps in the grid below the map set tree in Persephone is as follows:
- first show the chromosomes ordered by order_no assigned during loading the sequences.
- then show the rest of the maps, reversely ordered by size
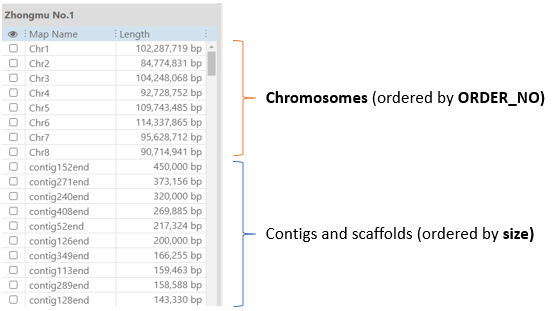
Mark some sequences as chromosomes - add chromosome records
It may happen that the decision to identify chromosomes comes after the sequences have been loaded. To add the chromosome records to an existing map set use the command edit chromosomes:
edit chromosomes <{mapSetId | path}>
for example, we decided to mark ChrUn as a chromosome entry:
PS> edit chromosomes "Oryza sativa japonica/Rice IRGSP-1"
Existing chromosomes for Oryza sativa japonica/Rice IRGSP-1:
CHROM_NAME CHROM_LENGTH MAP_NAME
Chr1 43,270,923 Chr1
Chr2 35,937,250 Chr2
Chr3 36,413,819 Chr3
Chr4 35,502,694 Chr4
Chr5 29,958,434 Chr5
Chr6 31,248,787 Chr6
Chr7 29,697,621 Chr7
Chr8 28,443,022 Chr8
Chr9 23,012,720 Chr9
Chr10 23,207,287 Chr10
Chr11 29,021,106 Chr11
Chr12 27,531,856 Chr12
There are 2 more maps that can be designated as chromosomes.
A - Add chromosomes, O - change order, R - remove chromosome flag (unmark), ESC - cancel: A
LINE_NO MAP_NAME CHROM_LENGTH
[ 0] ChrUn 633,585
[ 1] ChrSy 592,136
Type line number(s) of maps to be designated as chromosomes.
Use comma as a separator or use range of numbers like 0..10 : 0
ChrUn
Choose one of the options:
R - use regular expression to extract chromosome name from map name
F - (not implemented) use file with map and chromosome name pairs (one line for each pair, comma- or tab-delimited)
M - (not implemented) type the names manually
S - (not implemented) sort maps by name and use the order number
Your choice: R
Regular expression to extract chromosome name from the map name: (.+)
ChrUn ==> ChrUn
Do you want to insert the chromosome records listed above? (Y/N)Y
Inserted 1 chromosome record(s)
In the example above, first, a list of existing maps called chromosomes is displayed. The program found 2 maps that do not have an associated chromosome record. The first of them, ChrUn, listed under line_no 0 is selected.
In the current version, only one method of naming chromosomes is implemented: using a regular expression to extract the chromosome name from the map name. The program will search for the common prefix among selected maps and will suggest using it in the regular expression, so that the resultant chromosome name is as short as possible. The chromosome names are normally shown in the graphical representation of a genome, where they appear together with other chromosome names, so it is important to have them short.
The suggested regular expression (.+) will result in copying the map name into the chromosome name verbatim. If you want to shorten the name of the chromosome, you might want to remove the leading Chr by using the regular expression Chr(.+).
Reorder existing chromosomes
The order_no records of the sequences are considered only when sorting the chromosomes, as they are shown first, on top of the list of all maps. The rest of the sequences that are not chromosomes are ordered by size.
When loading the sequences, their order_no is assigned according to their order in the original FASTA file. It is not uncommon that the sequences in the file are given in the arbitrary order.
To change the chromosome order, run a command like
PS> edit chromosome "Oryza sativa/IRGSP-1.0.31"
This will list the current order of the sequences.
Existing chromosomes for Oryza sativa/IRGSP-1.0.31:
CHROM_NAME CHROM_LENGTH MAP_NAME ORDER_NO
9 23,012,720 9 49
10 23,207,287 10 50
12 27,531,856 12 51
8 28,443,022 8 52
11 29,021,106 11 53
7 29,697,621 7 54
5 29,958,434 5 55
6 31,248,787 6 56
4 35,502,694 4 57
2 35,937,250 2 58
3 36,413,819 3 59
1 43,270,923 1 60
There are several ways of reordering the maps. Most of the time, the chromosomes can be ordered by their "natural" order, which sorts the map names taking into account the numerical values that could be part of the names. This will ensure, for example, that Chr2 will precede Chr10. Note, that using a plain alpha-numeric sorting will put Chr2 after Chr10.
- To use the natural ordering, type 'N':
There are 49 more maps that can be designated as chromosomes.
A - Add chromosomes, O - change order,R - remove chromosome flag (unmark), ESC - cancel: O
N - natural order, F - use records from file, ESC - cancel: N
CHROM_NAME CHROM_LENGTH MAP_NAME ORDER_NO
1 43,270,923 1 0
2 35,937,250 2 1
3 36,413,819 3 2
4 35,502,694 4 3
5 29,958,434 5 4
6 31,248,787 6 5
7 29,697,621 7 6
8 28,443,022 8 7
9 23,012,720 9 8
10 23,207,287 10 9
11 29,021,106 11 10
12 27,531,856 12 11
Save the new order? (Y/N) Y
- An alternative way of reordering the chromosomes uses the records in a tab-delimited file, where a map name is followed by the order_no, such as:
Chr1 0
Chr2 1
...
Type 'F' at the prompt to select this mode of the ordering and provide the path to the file:
N - natural order, F - use records from file, M - order manually, ESC - cancel: F
Path to the file with chromosome order (mapName TAB orderNo):? /tmp/chrom-order.csv
CHROM_NAME CHROM_LENGTH MAP_NAME ORDER_NO
1 43,270,923 1 1
2 35,937,250 2 2
3 36,413,819 3 3
4 35,502,694 4 4
5 29,958,434 5 5
6 31,248,787 6 6
7 29,697,621 7 7
8 28,443,022 8 8
9 23,012,720 9 9
10 23,207,287 10 10
11 29,021,106 11 11
12 27,531,856 12 12
Save the new order? (Y/N) Y
Removing chromosome records
It is possible to remove the chromosome flag from maps, and mark them as non-chromosomes. If, for example, you would like to downgrade the map FLA1.3ch00, that currently has a status of a chromosome (called ch00), to the level of a scaffold, run this:
PS> edit chromosome "Solanum lycopersicum/FLA1.3"
13 existing chromosomes for Solanum lycopersicum/FLA1.3:
CHROM_NAME CHROM_LENGTH MAP_NAME ORDER_NO
ch00 5,490,904 FLA1.3ch00 0
ch01 95,309,210 FLA1.3ch01 1
ch02 52,158,778 FLA1.3ch02 2
ch03 66,828,682 FLA1.3ch03 3
ch04 67,650,907 FLA1.3ch04 4
ch05 66,930,101 FLA1.3ch05 5
ch06 46,398,775 FLA1.3ch06 6
ch07 69,121,753 FLA1.3ch07 7
ch08 63,731,143 FLA1.3ch08 8
ch09 67,978,353 FLA1.3ch09 9
ch10 68,636,165 FLA1.3ch10 10
ch11 56,952,951 FLA1.3ch11 11
ch12 68,816,593 FLA1.3ch12 12
O - change order, R - remove chromosome flag (unmark), ESC - cancel: R
LINE_NO CHROM_NAME CHROM_LENGTH MAP_NAME ORDER_NO
[0 ] ch00 5,490,904 FLA1.3ch00 0
[1 ] ch01 95,309,210 FLA1.3ch01 1
[2 ] ch02 52,158,778 FLA1.3ch02 2
[3 ] ch03 66,828,682 FLA1.3ch03 3
[4 ] ch04 67,650,907 FLA1.3ch04 4
[5 ] ch05 66,930,101 FLA1.3ch05 5
[6 ] ch06 46,398,775 FLA1.3ch06 6
[7 ] ch07 69,121,753 FLA1.3ch07 7
[8 ] ch08 63,731,143 FLA1.3ch08 8
[9 ] ch09 67,978,353 FLA1.3ch09 9
[10] ch10 68,636,165 FLA1.3ch10 10
[11] ch11 56,952,951 FLA1.3ch11 11
[12] ch12 68,816,593 FLA1.3ch12 12
Select line number(s) of maps to remove the chromosome flag.
Use comma as a separator or use range of numbers like 0..10 : 0
FLA1.3ch00
The following 1 chromosome record(s) will be cleared
CHROM_NAME CHROM_LENGTH MAP_NAME
ch00 5,490,904 FLA1.3ch00
Do you want to proceed? (Y/N) Y
DATA_VERSION updated
Deleted 1 chromosome record(s)
