Adding BED
The files in BED format can provide additional annotation for the sequence maps. The files can be dragged and dropped onto the main Persephone window, thus creating temporary tracks, or they can be added to the database via PersephoneShell's command 'add bed'.
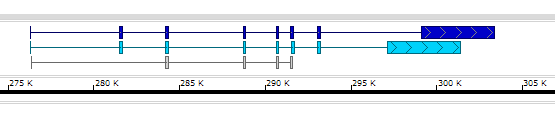
Here is an example of the BED file with 12 columns:
browser position chr1:253742-303353
browser hide all
track name="ItemRGBDemo" description="Item RGB demonstration" visibility=2 itemRgb="On"
chr1 253742 255091 teal_gene 0 + 253742 253742 250,200,100 2 104,155, 0,1194,
chr1 276321 303353 blue_gene 0 + 284187 299335 0,0,200 8 73,203,148,137,129,156,184,4257, 0,5160,7845,12411,14326,15177,16713,22775,
chr1 276323 291537 cyan_gene 0 + 276323 276323 0,210,250 8 73,203,148,137,129,156,184,4257, 0,5160,7845,12411,14326,15177,16713,20775,
chr1 276352 291629 nothing 0 + 284187 291629 0 5 42,148,137,129,131, 0,7814,12380,14295,15146,
Since the BED file uses a well-defined format, there is not much room for customization of the loading process. The [Bed] section will contain just a few instructions:
[Bed]
; BED specification: https://m.ensembl.org/info/website/upload/bed.html https://genome.ucsc.edu/FAQ/FAQformat.html#format1
; Sources (required): BED file(s) placed locally or accessible remotely via URL.
Sources="Samples/Bed/sample_exons.bed"
; SkipInvalidRecords: if true - skip line if it has invalid data (and print a warning), otherwise stop parsing and print an error.
;SkipInvalidRecords=true
; Commit frequency: indicates how often the process commits annotations. Every N annotations.; Commit frequency: indicates how often the process commits annotations. Every N annotations.
;CommitFrequency=1000
; RebuildSearchIndex: if true (default), the search index on Oracle will be rebuilt
; The setting has no effect for MySql.
; One of the reasons to set it to false is adding annotation in a batch, leaving the index rebuilding to the very last command
;RebuildSearchIndex=false
; Method (required): annotation method. If new, should be specified in METHOD section.
Method="BedSampleMethod"
; Track name (required): name of track
TrackName="Sample Bed track"
; TrackDescription: track description
TrackDescription="track with generic data"
; ParentGroupName: the new track will be placed under a parent node with this name.
; To reduce the number of track nodes on the top level, group the tracks of similar type.
;ParentGroupName=bed
; IsShownFirst: if false, the track will not be shown by default when the map is opened for the first time
IsShownFirst=false
; Assume that 'thick' columns (index 7 and 8) define CDS range for annotation, if false, 'thick' columns will be ignored. true by default
;AssumeThickRangeIsCds=false
According to BED specification, the first three columns of the BED file are required. The other columns are optional but all rows should have the same number of columns. The missing values should be denoted as a dot (.) symbol. As the map names sometimes may contain spaces, parsing the data rows will expect only the TAB symbol as the delimiter. The first (optional) lines beginning with 'browser' or 'track' is allowed to contain the space as the delimiter.
The score column (index=4) values will be ignored by Persephone. If you need to specify the color for each feature, put the R,G,B values in the column with index 8 (all indexes are 0-based).
The [Bed] section contains the usual information necessary to create tracks, such as Sources, TrackName or TrackDescription. In addition to that, you can specify a parent track node (ParentGroupName) used to group the tracks of similar type. This is recommended in case when a map contains many tracks; such grouping will reduce the number of top-level nodes of the track tree shown on each map.
The columns "thickStart" and "thickEnd" (index=6 and 7)
chr1 253742 255091 teal_gene 0 + 253742 253742 250,200,100 2 104,155, 0,1194,
chr1 276321 303353 blue_gene 0 + 284187 299335 0,0,200 8 73,203,148,137,129,156,184,4257, 0,5160,7845,12411,14326,15177,16713,22775,
chr1 276323 291537 cyan_gene 0 + 276323 276323 0,210,250 8 73,203,148,137,129,156,184,4257, 0,5160,7845,12411,14326,15177,16713,20775,
chr1 276352 291629 nothing 0 + 284187 291629 0 5 42,148,137,129,131, 0,7814,12380,14295,15146,
The records in the bed file can be used to provide the gene models. If you know that the thickStart and thickEnd designate the boundaries of the CDS, set AssumeThickRangeIsCds to true.
Running the command
The new tracks from the BED files are created using the command 'add bed'.
Test mode:
add bed -c controlBed.ini -t
or, the actual loading with verbose output:
add bed -c controlBed.ini -v
