Adding Synteny Ribbons
When showing syntenic maps on one screen, Persephone links them by connecting identical markers or orthologous genes. There is one more type of connectors that visualizes homology between the regions: synteny "ribbons":
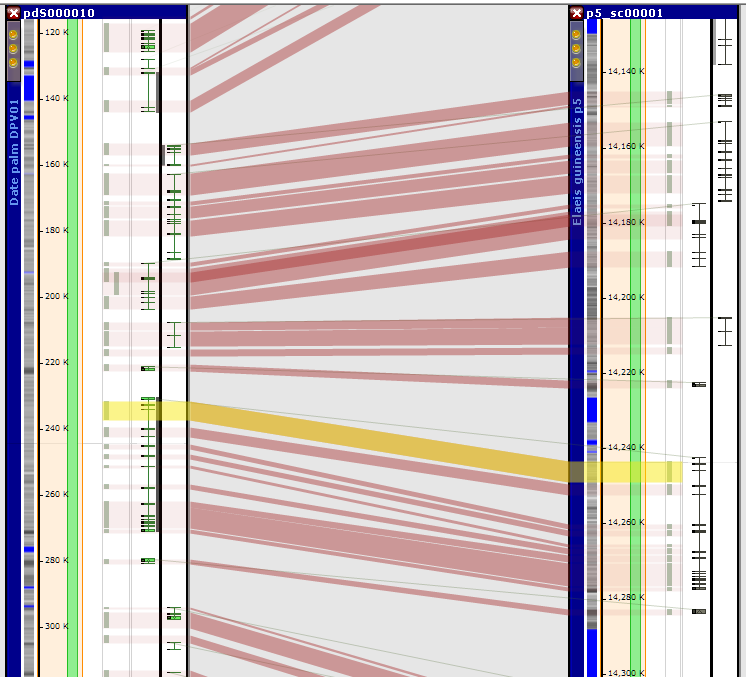
Each ribbon shows which regions of the two maps are homologous. To define a ribbon you will need the map names with coordinates of start and end of the regions. This information can be easily provided in a tab-delimited file where each ribbon data is stored on a separate line:
pdS0000010 60215184 60215189 p5_sc0001 63490876 63490881
pdS0000010 60215190 60215212 p5_sc0001 63490883 63490905
pdS0000010 60215213 60215242 p5_sc0001 63490907 63490936
Each ribbon can contain a qualifier, such as the score of the match.
Loading this information is done by the command 'add ribbons'.
Test mode (just testing):
add ribbons -c control.ini -t
Verbose mode (loading to the database):
add ribbons -c control.ini -v
Loading ribbons from a text file
A sample INI file for loading the ribbons from the text file is below:
[ProcessRun]
; RunDescription: if specified, a custom description will be used,
; otherwise, "Added synteny between {TargetMapSet Accession No.} and {QueryMapSet Accession No.} from {Source}." will be used.
RunDescription="Added test syntenic regions between tomato and potato"
[TargetMapSet]
; Either MapSetId or MapSetPath is required.
; MapSetId: id of a target map set.
;MapSetId=242685112
; MapSetPath: path of a target map set.
MapSetPath="Solanum lycopersicum/SL4.0"
[TargetMapMapping]
; If no mapping is found in this section, psh assumes that each MAP_NAME in file exactly matches a MAP_NAME in DB.
; If map names in file are different from those in DB, map each MAP_NAME in file to its MAP_NAME in DB.
; Otherwise, no syntenic region will be loaded.
;MAP_NAME in file=MAP_NAME in DB
;chr1=CHR1
; MapsIdentifiedBy: if all maps in the file instead of the map name are identified by their alternative IDs like MAP_ID, ACCESSION_NO or GENOME_DNA_ID,
; provide the mapping with just one line using either MapName, MapId, AccessionNo or GenomeDnaId
;MapsIdentifiedBy=GenomeDnaId
[QueryMapSet]
MapSetPath="Solanum tuberosum/DM_v4.03"
; Same logic for map mapping as described above
[QueryMapMapping]
; If no mapping is found in this section, psh assumes that each MAP_NAME in file exactly matches a MAP_NAME in DB.
; If map names in file are different from those in DB, map each MAP_NAME in file to its MAP_NAME in DB.
; Otherwise, no syntenic region will be loaded.
;MAP_NAME in file=MAP_NAME in DB
;chr1=CHR1
; MapsIdentifiedBy: if all maps in the file instead of the map name are identified by their alternative IDs like MAP_ID, ACCESSION_NO or GENOME_DNA_ID,
; provide the mapping with just one line using either MapName, MapId, AccessionNo or GenomeDnaId
;MapsIdentifiedBy=GenomeDnaId
[Synteny]
; Source (required): a chain or a GFF file or a TXT file placed locally or remotely accessible via URL.
Source=$DATA/ribbons.txt
; CoordinateSystem: 1 (one-based) / 0 (zero-based). Default value is 1.
; Chain is usually 0-based, while Gff is 1-based.
CoordinateSystem=1
; Commit frequency: indicates how often the process commits markers. Every N markers.
CommitFrequency=10000
; FileType (required): {Chain|Gff|Text}
FileType=Text
;---------------------------------------------------------------------------------------------------------------
; Parsing Information
;---------------------------------------------------------------------------------------------------------------
; SkipHeaderLines: the number of lines to skip parsing
;TextSkipHeaderLines=0
; CommentPrefix: comment prefix to skip parsing
;TextCommentPrefix="#"
; Delimiter: specify one among Colon(:), Comma(,), Period(.), Hyphen(-), SemiColon(;), Slash(/), Tab(\t), VerticalBar(|)
TextDelimiter=Tab
;-------------------------
; Sequence alignment programs search subjects using query sequences.
; We assume that the alignment results must contain information as below.
; - subject (target) coordinates: mapName , start, end, (strand)
; - query coordinates: mapName , start, end, (strand)
; - ribbon color
; Index: column index(0-based)
;TextSyntenyNameIndex=0
TextTargetMapNameIndex=3
TextTargetStartIndex=4
TextTargetEndIndex=5
;TextTargetStrandIndex=0
TextQueryMapNameIndex=0
TextQueryStartIndex=1
TextQueryEndIndex=2
;TextQueryStrandIndex=0
;TextRibbonColorIndex=8
; TextQualifierIndex: Text column whose value contains synteny qualifiers.
;TextQualifierIndex.Index=qualifierName((:displayText),dataType,dataFormat)
;TextQualifierIndex.1="Score"
[DbSequences]
; The ID columns below are used in loading synteny data.
; If there is no sequence/trigger assigned to these columns, you must specify a sequence for them.
;TABLE_NAME.COLUMN_NAME=SEQUENCE_NAME
;PROCESS_RUN.RUN_ID=ID_SEQ
;TRACK_CONNECTOR.TRACK_CONNECTOR_ID=ID_SEQ
;TRACK_CONNECTOR_QUALIFIER.QUALIFIER_ID=ID_SEQ
Note
The current version of Persephone (4.3.0.127) has practical limitations on the number of simultaneously displayed ribbon connectors. It works reasonably well when the number of ribbons is less than 1,000. Otherwise, you might experience performance issues.
Loading synteny ribbons from chain files
If the information about related regions is provided in a form of chain file, you will have a few new options.
The chain file contains records of two types: the chain boundaries (bold font below) showing on a larger scale, which region is similar to which, and the fine structure of each chain listing insertions/deletions on both maps:
chain 114691 chr1 308452471 + 124446006 124447236 Chr1 307041717 + 125685125 125686356 398654
754 0 1
476
chain 107215 chr1 308452471 + 124447236 124457416 Chr3 235667834 + 167570846 167580982 245115
225 17 14
194 1 0
146 34 36
68 163 149
280 1 0
3042 85 84
2598 46 46
399 1734 1736
137 107 103
135 17 4
201 34 36
68 162 149
286
By default, PersephoneShell will read only the chain records, ignoring the fine structure. The parameter IgnoreChainsSmallerThan will control which chains will be loaded and which ones will be skipped. If the parameter is not used, all chains will be considered.
If you think that adding the ribbons based on the fine structure will not overwhelm the graphics, you can set LoadChainFineStructure variable to true. In that case, the ribbons will be formed by the records of the fine structure and not by the chains themselves.
To control the resolution of the ribbons, use parameter IgnoreGapsSmallerThan - the ribbons separated by small gaps will be merged together. This helps reducing the number of ribbons to be stored and displayed, lowering the stress on the system.
[Synteny]
; Source (required): a chain or GFF file located locally or remotely accessible via URL.
Source="http://hgdownload.cse.ucsc.edu/goldenPath/hg19/vsMm10/hg19.mm10.all.chain.gz"
; Number format culture: specifies a culture name used to parse numbers in data. Default value is en - English.
; e.g. de - German, es - Spanish, fr - French. For more cultures, https://msdn.microsoft.com/en-us/goglobal/bb896001.aspx
;NumberFormatCulture="fr"
; CoordinateSystem: 1 (one-based) / 0 (zero-based). Default value is 1.
; Chain is usually 0-based, while Gff 1-based.
CoordinateSystem=0
; Commit frequency: indicates how often the process commits markers. Every N markers.
CommitFrequency=10000
; FileType (required): {Chain|Gff|Text}
FileType=Chain
; IgnoreChainsSmallerThan: Filter the chains. The chains smaller than this size (in bp) will be ignored.
IgnoreChainsSmallerThan=50000
; IgnoreGapsSmallerThan: ignore small gaps in the chain internal structure.
IgnoreGapsSmallerThan=3000
; LoadChainFineStructure: if true, load the chain's fine structure listed in the lines following the chain info.
; If false, the synteny ribbons will be based purely on the chain records, the fine structure will be ignored (default:false)
LoadChainFineStructure=true
When using the chain records only, you might find helpful a histogram of the chain size distribution. It will estimate the number of ribbon elements to be loaded at given threshold (IgnoreChainsSmallerThan) value:
Estimates of threshold (IgnoreChainsSmallerThan) and the number of ribbons that would be loaded with this value of IgnoreGapsSmallerThan:
67,108,864 9 ribbons
33,554,432 26 ribbons
16,777,216 50 ribbons
8,388,608 92 ribbons
4,194,304 137 ribbons
2,097,152 205 ribbons
1,048,576 252 ribbons
524,288 295 ribbons
262,144 359 ribbons
131,072 510 ribbons
65,536 908 ribbons
32,768 2,246 ribbons
16,384 5,983 ribbons
8,192 16,436 ribbons
4,096 42,931 ribbons
2,048 129,091 ribbons
1,024 469,223 ribbons
512 1,805,515 ribbons
256 2,537,247 ribbons
128 3,179,870 ribbons
64 3,376,049 ribbons
32 3,392,421 ribbons
Depending on the total number of map pairs, decide which value of IgnoreChainsSmallerThan will result in the number of ribbons that does not exceed a dozen of thousand ribbons per pair of maps. With the larger counts, you risk having performance issues.
If LoadChainFineStructure is true, each chain will be split into multiple small ribbons. Some of them can be merged, based on IgnoreGapsSmallerThan parameter. If both gaps (query and target) are smaller than the specified value, the gap will be ignored and the neighboring ribbons will be merged. A matrix for different values of IgnoreChainsSmallerThan and IgnoreGapsSmallerThan will print the number of ribbons that would pass the filter. This should help you choosing the right pair of parameters.
- 176,792 ribbons will be loaded. IgnoreChainsSmallerThan=65,536, IgnoreGapsSmallerThan=3,000
Estimates of threshold (IgnoreChainsSmallerThan) and the number of ribbons that would be loaded with various values of [IgnoreGapsSmallerThan]:
[0] [1] [2] [5] [10] [20] [50] [100] [500] [1,000] [3,000]
67,108,864 4,919,631 4,919,622 3,476,105 1,879,897 1,029,398 527,434 297,352 250,179 105,763 66,329 28,386
33,554,432 14,373,807 14,373,781 10,154,805 5,496,384 2,994,053 1,512,642 838,886 704,648 296,902 183,704 75,519
16,777,216 20,191,903 20,191,853 14,262,446 7,726,029 4,210,498 2,126,646 1,178,233 990,834 417,006 257,636 105,683
8,388,608 25,826,043 25,825,951 18,254,912 9,913,853 5,415,754 2,734,148 1,511,162 1,272,086 534,186 328,988 134,412
4,194,304 29,011,994 29,011,857 20,510,957 11,140,215 6,084,793 3,070,356 1,696,982 1,429,543 602,750 372,141 152,334
2,097,152 31,129,984 31,129,779 22,036,744 11,990,234 6,559,375 3,307,141 1,821,890 1,534,410 648,222 400,415 164,224
1,048,576 31,798,760 31,798,508 22,519,629 12,260,057 6,710,692 3,384,290 1,864,477 1,570,821 664,477 410,673 168,490
524,288 32,094,112 32,093,817 22,732,363 12,378,779 6,777,514 3,418,284 1,883,355 1,586,890 671,792 415,372 170,583
262,144 32,314,603 32,314,244 22,891,580 12,467,693 6,827,377 3,443,936 1,897,819 1,599,217 677,052 418,571 172,005
131,072 32,519,854 32,519,344 23,039,313 12,548,505 6,872,074 3,467,236 1,911,529 1,611,066 682,859 422,471 173,969
65,536 32,773,084 32,772,176 23,222,154 12,649,496 6,928,203 3,497,033 1,929,704 1,626,935 691,252 428,353 176,792
32,768 33,162,015 33,159,769 23,503,031 12,803,694 7,014,947 3,544,201 1,959,721 1,653,507 707,237 440,327 182,732
16,384 33,619,742 33,613,759 23,835,535 12,989,790 7,123,874 3,609,676 2,005,488 1,694,793 735,265 462,381 193,250
8,192 34,273,903 34,257,467 24,311,814 13,261,101 7,285,787 3,712,070 2,082,028 1,764,932 784,847 502,252 213,144
4,096 35,265,694 35,222,763 25,035,254 13,665,037 7,528,954 3,871,798 2,206,482 1,878,483 865,225 566,001 251,453
2,048 37,653,208 37,524,117 26,787,690 14,637,840 8,077,993 4,248,234 2,510,723 2,145,362 1,031,338 696,961 339,898
1,024 43,915,656 43,446,433 31,219,150 17,036,330 9,441,119 5,191,675 3,282,592 2,794,060 1,448,280 1,054,469 680,030
512 61,504,971 59,699,456 43,890,442 23,447,388 12,518,865 7,176,131 5,018,620 4,366,400 2,800,166 2,390,761 2,016,322
256 65,977,920 63,440,673 46,749,596 25,111,540 13,627,033 8,070,236 5,834,736 5,131,229 3,531,898 3,122,493 2,748,054
128 68,313,229 65,133,359 48,117,315 26,034,287 14,357,347 8,737,944 6,484,469 5,774,291 4,174,521 3,765,116 3,390,677
64 68,820,443 65,444,394 48,389,295 26,254,630 14,559,948 8,935,222 6,680,660 5,970,470 4,370,700 3,961,295 3,586,856
32 68,854,665 65,462,244 48,406,469 26,271,136 14,576,324 8,951,594 6,697,032 5,986,842 4,387,072 3,977,667 3,603,228
If needed, delete the set of ribbons by the command delete run, using RunId of the corresponding job. To find RunId, list the jobs with the type of the command used when loading (add ribbon):
list run -T ribbon
Loading synteny ribbons from gff files
The typical purpose of gff file is to provide location of features on maps that belong to one map set. The synteny ribbons connect two intervals that reference two different maps. So, the line in gff file should contain both sets of coordinates - for the query interval on one map and for the target region on the other.
##gff-version 3
Vu01 DAGchainer syntenic_region 64390 1882809 4545.0 - . Name=Gm06;matches=Gm06:49767515..51299643;median_Ks=0.3641
Vu01 DAGchainer syntenic_region 64390 260549 402.0 + . Name=Gm04;matches=Gm04:52243031..52358762;median_Ks=0.4196
Vu01 DAGchainer syntenic_region 230234 249905 200.0 + . Name=Gm04;matches=Gm04:52200630..52225676;median_Ks=0.3752
Vu01 DAGchainer syntenic_region 298633 332448 185.0 - . Name=Gm04;matches=Gm04:12143112..12222024;median_Ks=0.3257
Vu01 DAGchainer syntenic_region 967088 1088349 340.0 + . Name=Gm04;matches=Gm04:14122926..14885446;median_Ks=0.4443
Vu01 DAGchainer syntenic_region 1204172 1310597 118.0 - . Name=Gm04;matches=Gm04:17927529..18391402;median_Ks=0.4079
In the example above, the coordinates of the target location of the match are given in the standard gff columns for map name (column 1), start (column4), end (column 5) and strand (column 6). The location of the query is provided in one of the attributes: matches=Gm06:49767515..51299643. It is likely that the format of the query coordinates will be different in different sources, so, to provide some flexibility, PersephoneShell will accept a QueryFormat string that denotes the map name, start, end and, optionally, strand as {MapName},{Start},{End} and {Strand} respectively. Put them in the same format as appears in the value of the corresponding attribute. For example, to correctly parse the query region written as Gm06:49767515..51299643, use QueryFormat="{MapName}\:{Start}\.\.{End}":
[Synteny]
; Source (required): a chain or GFF file located locally or remotely accessible via URL.
Source=$DATA/cowbean/vigun.IT97K-499-35.gnm1.ann1.x.glyma.Wm82.gnm2.ann1.gff3
; CoordinateSystem: 1 (one-based) / 0 (zero-based). Default value is 1.
; Chain is usually 0-based, while Gff 1-based.
CoordinateSystem=1
; Commit frequency: indicates how often the process commits markers. Every N markers.
CommitFrequency=10000
; FileType (required): {Chain|Gff|Text}
FileType=Gff
;---------------------------------------------------------------------------------------------------------------
; Parsing Information
;---------------------------------------------------------------------------------------------------------------
; GffSources: Gff column 2. Database name or software that generated these features.
; if not specified, all the sources will be included.
;GffSources=""
;------------------------
; GffTypes: A hit is a region of sequence, aligned to another sequence with some statistical significance.
; if not specified, all the GFF parent types will be included.
;GffTypes="match","match_set"
;------------------------
; Sequence alignment programs search subjects using query sequences.
; In an alignment output in GFF3, we assume that each sequence coordinate information is provided as below.
; - subject coordinates: seqid (GFF column 1), start (GFF column 4), end (GFF column 5), strand (GFF column 7)
; - query coordinates: in attributes
;
; A query coordinate can be given as either a single attribute of formatted string or multiple attributes.
; 1) single formatted attribute
; e.g. Target="C1 2035 2977 -"
; GffQueryAttributeKey: Gff attribute key whose value contains formatted string for query coordinate.
GffQueryAttributeKey="matches"
; QueryFormat: a formatted string
; {MapName} : query map name
; {Start} : query start position
; {End} : query end position
; {Strand}: query strand +/-/.
QueryFormat="{MapName}\:{Start}\.\.{End}"
; 2) multiple attributes
; e.g. QueryMapName=C1;QueryStart=2035;QueryEnd=2977;QueryStrand=-
;GffQueryMapNameAttributeKey="QueryMapName"
;GffQueryStartAttributeKey="QueryStart"
;GffQueryEndAttributeKey="QueryEnd"
;GffQueryStrandAttributeKey="QueryStrand"
;-------------------------------------
; e.g. ID=A01.match.57fe0a8b0728bc00;percent_identity=83.66219
; GffQualifierAttributeKey: Gff attribute key whose value contains synteny qualifiers.
;GffQualifierAttributeKey.AttributeKey=qualifierName((:displayText),dataType,dataFormat)
;GffQualifierAttributeKey.percent_identity=PercentIdentity
