Adding Spliced Alignments (SAM)
Persephone can show gene models that result from spliced alignment of transcripts, such as full- or partial-length mRNA/cDNA or ESTs:
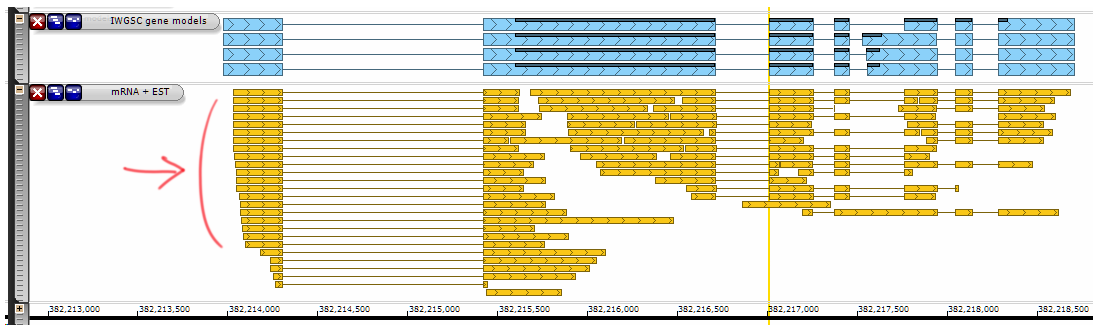
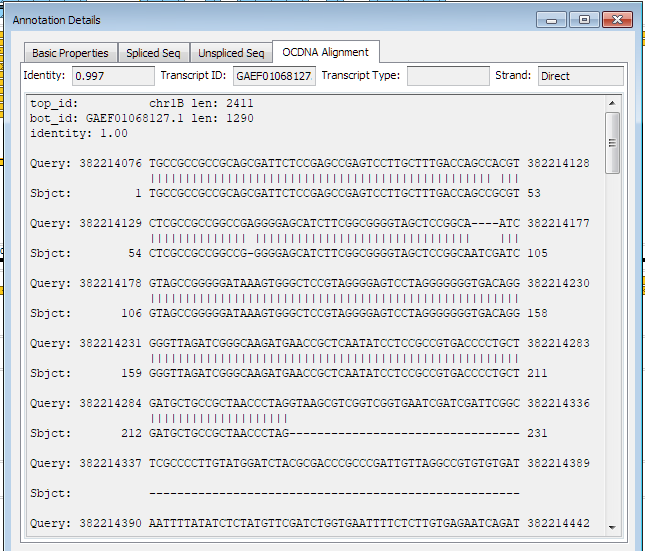
In case the gene model is produced by spliced alignment, a new tab will appear that would show the alignment on the level of base pairs:
Loading the spliced alignments provided as SAM files is done by the command add sam, for example:
add sam -c sam.ini -t
The format of the INI file is somewhat similar to the one for loading the annotation, but is simpler:
[ProcessRun]
; Run description: if specified, a custom description will be used,
; otherwise, "Added spliced alignments for {MapSet AccessionNo} from {Source}." will be used.
RunDescription="Load mRNA spliced alignment annotation for wheat. mRNA loaded from NCBI"
[MapSet]
; Either MapSetId or MapSetPath is required.
; MapSetId: id of a target map set.
;MapSetId=29
; MapSetPath: path of a target map set.
MapSetPath="/Triticum aestivum/Wheat IWGSCv1.0"
[Method]
; To add a new annotation method, specify a method name, CDS flag and a color to be shown in the annotation track.
; You don't need to provide this info if the method already exists in the database.
; The CDS flag is a boolean value that indicates if the exons predicted by this method represent the CDS part of the gene only.
; Most of the time, for the spliced alignments, the CDS flag is 'false'.
; Supported color name is available in http://www.flounder.com/csharp_color_table.htm
; If a method already exists, it will be updated. Otherwise, a new method will be added.
;Name=IsCds,{NamedColor|HTML hex code|R,G,B}
HISAT2=false,250,180,50
[Annotation]
; Provide the path or URL to the source SAM file (typically produced by a spliced aligner such as 'hisat2' or 'Magic-BLAST')
; Note: a single file is supported only
Sources=r:\wheat\wheatMrna.sam
; Commit frequency: indicates how often the process commits annotations. Every N annotations.
CommitFrequency=1000
; Method (required): annotation method. If new, should be specified in METHOD section above.
Method="HISAT2"
; Track name (required): name of track
TrackName="mRNA+EST"
; TrackDescription: track description
TrackDescription="Wheat mRNA and EST (1.6 M entries) from NCBI were splice-aligned using hisat2"
; RebuildIndex: indicates if qualifier indices in Oracle database should be rebuilt or not. The default value is 'true'.
; To save time on indexing, RebuildIndex can be set to 'false' if you know that there will be another loading following this one.
; In that case, the indexing will be done only once, after all annotations have been added.
;RebuildIndex=false
[QualifierLinks]
; The ID of Each aligned transcript will be stored as qualifier "Transcript"
; This is the only qualifier for the splice-aligned models
; %s in the link is where a Transcript name is placed.
;Transcript=PLACEHOLDER_URL
Transcript="https://www.ncbi.nlm.nih.gov/nuccore/%s"
[MapMapping]
; If no mapping is found in this section, psh assumes that each MAP_NAME in file exactly matches a MAP_NAME in DB.
; If map names in file are different from those in DB, map each MAP_NAME in the file to a MAP_NAME in DB.
; The manual mapping is below:
;MAP_NAME_IN_FILE=MAP_NAME in DB
;Chr1=Chr.1
;Chr2=Chr.2
;Chr3=Chr.3
; Check 'printmapping' command that may help generating the name mapping tables.
; Alternatively, use MapIdentifiedBy
; MapsIdentifiedBy: if all maps in the file instead of the map name are identified by their alternative IDs like MAP_ID, ACCESSION_NO or GENOME_DNA_ID,
; provide the mapping with just one line using either MapName, MapId, AccessionNo or GenomeDnaId, for example:
; MapsIdentifiedBy: one of MapId, AccessionNo, GenomeDnaId, MapName (default)
;MapsIdentifiedBy=AccessionNo
; LoadListedMapsOnly: if true, only data for the maps listed in this section will be added.
; If false, PersephoneShell will still try to match names from the file to maps in the database
; using MAP_NAME, MAP_ID or ACCESSION_NO, and if the map is not found, the data line will be skipped
;LoadListedMapsOnly=true
;MAP_NAME_IN_FILE=MAP_NAME_IN_DB
[DbSequences]
; Oracle only
; The ID columns below are used in loading annotations.
; If there is no sequence/trigger assigned to these columns, you must specify a sequence for them.
;PROCESS_RUN.RUN_ID=ID_SEQ
;GDNA_ANNOT.ANNOT_ID=ID_SEQ
;DESCRIPTION.DESCR_ID=ID_SEQ
;TRACK.TRACK_ID=ID_SEQ
Loading of the SAM file will create an annotation track where the gene models will have a predefined qualifier 'Transcript'. It can be used to generate a hyper-link to an external source of the transcript, such as NCBI (see QualifierLinks section above).
