Run BLAST
BLAST (Basic Local Alignment Search Tool) is a set of widely-used sequence comparison tools. Persephone can run the following set of BLAST programs. Click the BLAST program name in the Program column for steps to run the program.
|
Program |
Query |
Database |
|
nucleotide |
nucleotide |
|
|
protein |
protein |
|
|
nucleotide (translated in all six frames by the program) |
protein |
|
|
protein |
translated nucleotide (all six frames) |
To run a BLAST comparison, click the Run Blast button ( ) in the toolbar in the main screen.
) in the toolbar in the main screen.
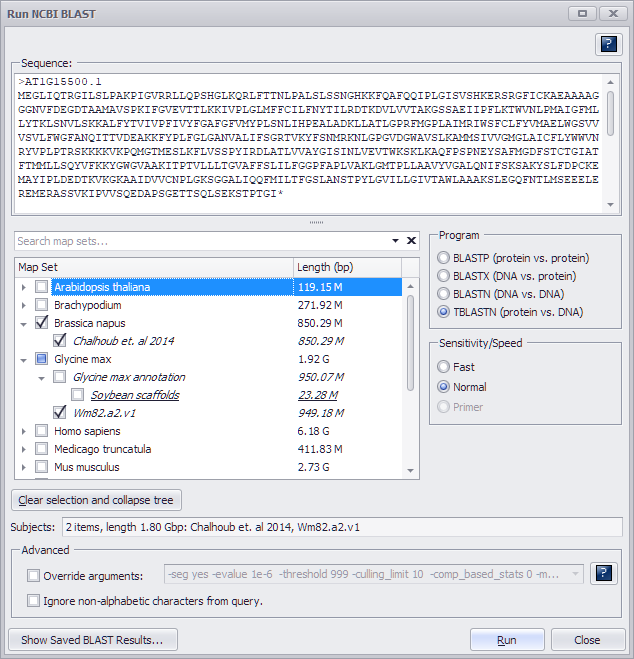
Enter the query sequence in the text box in FASTA format. If you are using a single sequence query, you do not have to include a definition line (i.e., ">" followed by text uniquely identifying the sequence) but it is good practice to always include one. If the query contains more than one sequence, however, each query sequence must have a unique identifier in the FASTA description line.
Select a target genome (or genomes) and choose a Sensitivity/Speed listed below.
- Fast. This option is less sensitive but will detect nearly identical matches.
- Normal. The Normal option has better sensitivity by relaxing the threshold on evaluation.
- Primer. The Primer option is used for BLASTing short sequence lengths and is only available for BLASTN and TBLASTN. To find a match for Primer the threshold on evaluation needs to be increased to 100 or even 1000 by entering "-evalue 100" or "-evalue 1000", respectively, in the Override arguments text box (described below). Otherwise, the hits for short sequences will be filtered by default BLAST settings.
Note:
The query sequence can be provided by the system or from the public (or other outside) source.
If you want to change the preset arguments of BLAST, click the Override arguments checkbox in the Advanced panel and modify the arguments in the text box. The help button to the right of the text entry box will produce a pop-up window with a detailed explanation of the arguments for the BLAST programs.
If the sequences contain non-alphabetic characters such as digits or brackets, check the Ignore non-alphabetic characters from query checkbox. The program will make sure that the query sequence corresponds to the expected type (DNA or protein) used by the selected type of BLAST.
Tip
For additional NCBI information see the NCBI BLAST Command Line Applications User Manual.
The following shows a valid entry for BLASTP.

Note, that as the subject data set, BLASTP will use a collection of proteins generated from the gene models predicted by selected method. If a map set has several gene model tracks predicted by different methods, under the map set node of the tree you will see several check boxes - one for each of the methods.
Click the OK button to run the BLAST comparison. Clicking the Cancel button will exit the BLAST tool. Note, that if you click the Show Previous BLAST Result button the last BLAST results you obtained will be displayed in the search results window, discussed below. The following shows a sample output for the BLASTP program.
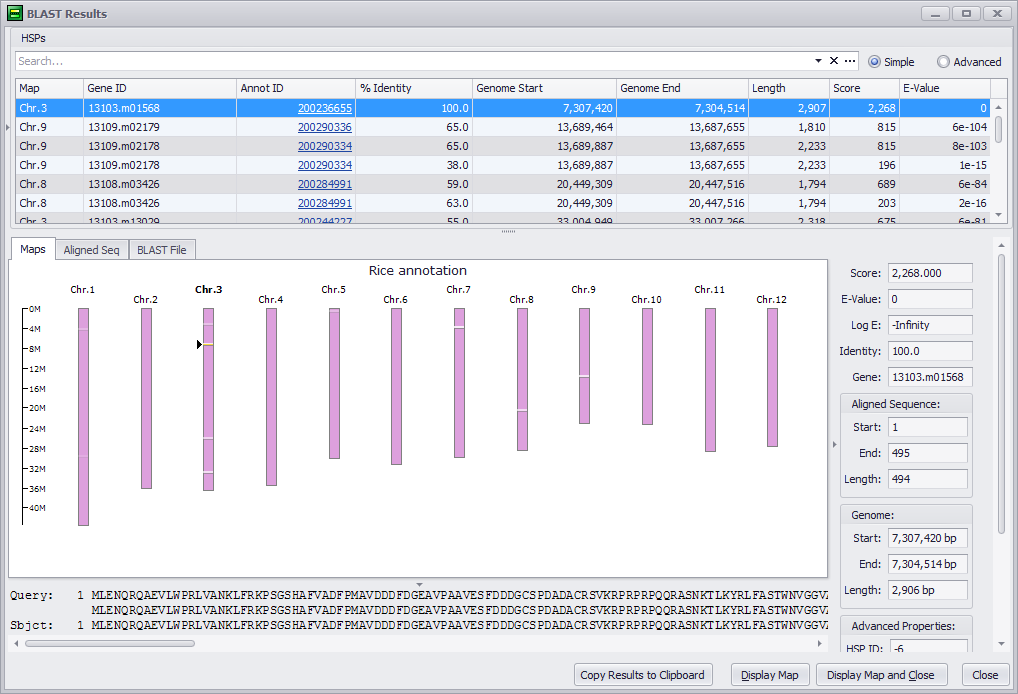
The data displayed by the BLAST Results window is described in BLAST Results Window. Click the following links for step-by-step instructions for each BLAST program:
Tip
See Mapping Primers for PCR Products for guidelines on using Persephone's real-time BLAST tool to search for primer hits.
