Web Persephone: Import variants
Sample files: https://persephonesoft.com/data/import/SNPs_lifted_final2_sorted_v1x.vcf.gz + https://persephonesoft.com/data/import/SNPs_lifted_final2_sorted_v1x.vcf.gz.csi
You can import files containing variants (e.g. SNPs or insertions/deletions) in VCF format, accompanied by a TBI or CSI index file.
Note
Currently, variants may be imported only into your local browser session, and are not preserved in long-term private storage.
The index file is required for VCF files larger than 15 MB:
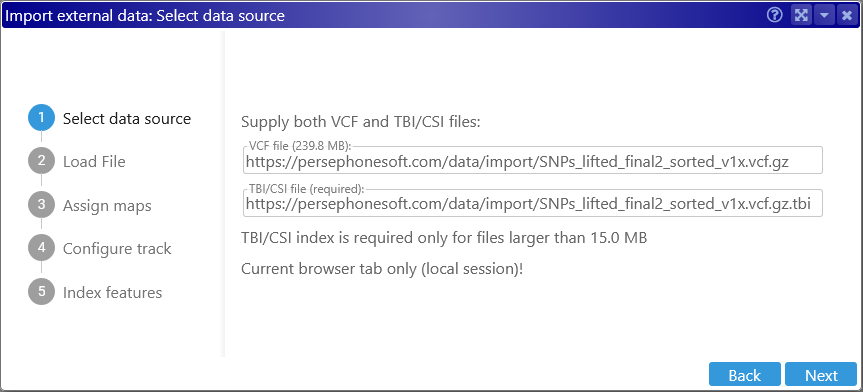
By default, Persephone automatically suggests a TBI index file, but you can always provide a different URL; in this example, the index is a CSI file: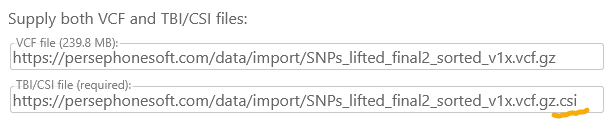
After the file is fully loaded, you will need to assign it to the correct map set, as usual. By default, maps are matched based on map names, but you may also choose to match them based on map accession:
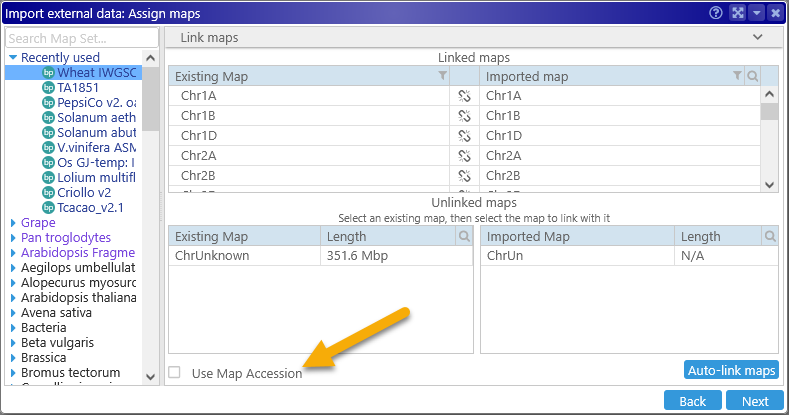
This can be useful when loading VCF files from external sources such as NCBI, whose data files typically use map accessions rather than human-readable names.
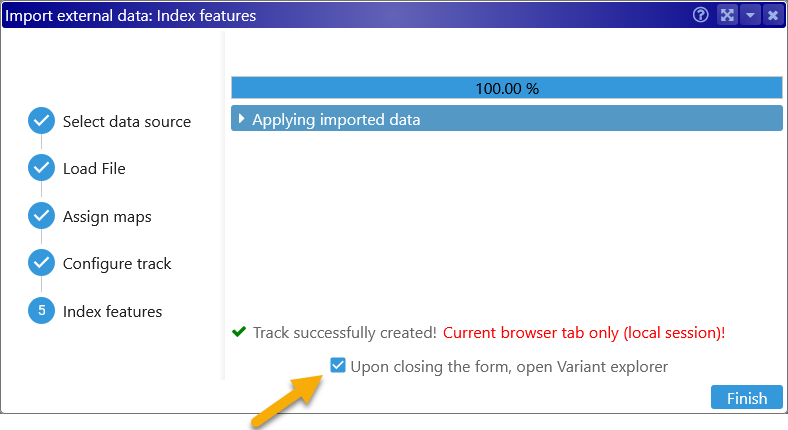
Click Next once again to finish importing the file; you may optionally choose to immediately view the loaded samples in the Variant Explorer:
The newly imported samples will be highlighted in light pink, both in the Variant Explorer and on the Variants track, indicating that they exist only in the local browser session, and will disappear when you reload Persephone:
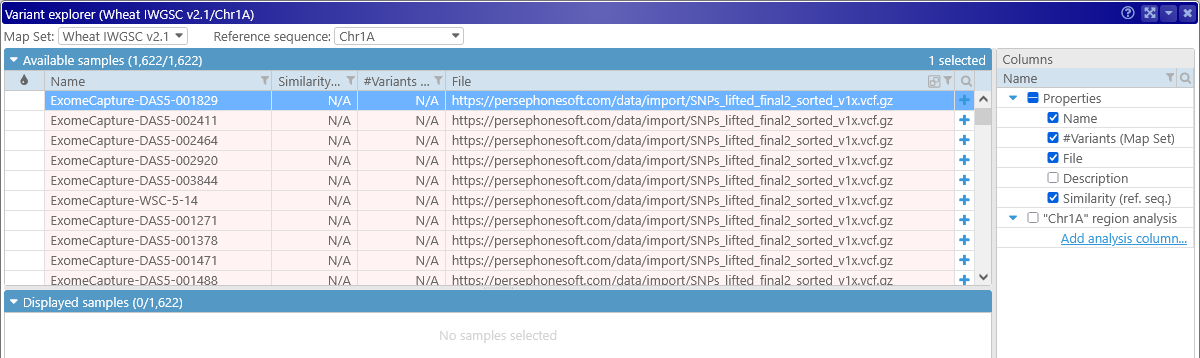
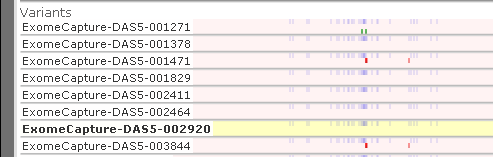
In addition, these samples will be missing some statistical information, such as their similarity to the reference sequence and the total number of variants per sample. Because of this, their histograms will be rendered in gray (and in low resolution):
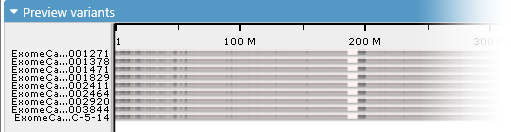
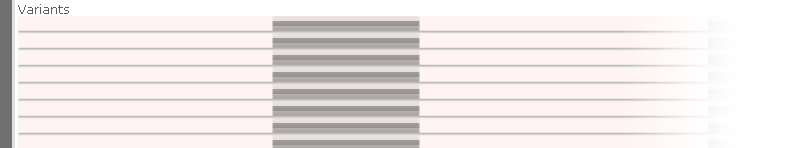
This happens because the TBI/CSI index files accompanying the imported VCF file provide only a coarse overview of the variants contained within it. If you wish to use the full set of Persephone's features when browsing the imported samples, you could load them into Persephone's permanent database using PersephoneShell.
