Web Persephone: Import synteny ribbons
Sample files: https://persephonesoft.com/data/import/coffee-chr-2.paf, https://persephonesoft.com/data/import/coffee-chr-2.ribbon
You can import syntenic blocks between a pair of map sets, from files in PAF format (as generated by minimap2) or Persephone's own Ribbon format (described below). In both cases, Persephone generates a Synteny track based on the loaded alignments.
Note
Currently, syntenic blocks may be loaded only into your local browser session, and are not preserved in long-term private storage.
Loading a PAF file
PAF files are most often generated by the minimap2 sequence alignment tool. For example, you could export the 2nd chromosome from two different varieties of Coffee (Cara_1.0 / chromosome 2c and C. canephora DH200-94 / chr2), then run a command like this:
minimap2 Cara_1.0_chromosome_2c.fa NCBI_GCA_900059795.1_chr2.fa -o coffee-chr-2.paf -w 100 -N2 -k 28 --secondary "no" -n 50
Doing so will generate a file similar to this one:
chr2 54522928 1913458 3668220 + chromosome_2c 66155350 1933959 3726389 542823 1899924 60 tp:A:P cm:i:21967 s1:i:484204 s2:i:5431 dv:f:0.0021 rl:i:43026
chr2 54522928 383369 1745800 + chromosome_2c 66155350 415962 1787337 411096 1469501 60 tp:A:P cm:i:16531 s1:i:363527 s2:i:1277 dv:f:0.0023 rl:i:43026
chr2 54522928 16899161 17755896 + chromosome_2c 66155350 16071734 16937662 281264 914874 60 tp:A:P cm:i:11451 s1:i:256306 s2:i:0 dv:f:0.0011 rl:i:43026
chr2 54522928 18988693 19720073 + chromosome_2c 66155350 17813710 18578900 243294 800925 60 tp:A:P cm:i:9867 s1:i:218920 s2:i:5131 dv:f:0.0014 rl:i:43026
chr2 54522928 11171307 11891640 + chromosome_2c 66155350 11049418 11811594 239832 792063 60 tp:A:P cm:i:9710 s1:i:216279 s2:i:41112 dv:f:0.0020 rl:i:43026
chr2 54522928 3895663 4532476 + chromosome_2c 66155350 4026534 4650758 193204 667862 60 tp:A:P cm:i:7779 s1:i:175667 s2:i:0 dv:f:0.0019 rl:i:43026
...
Drag-and-drop the output file onto Persephone's browser tab (or open the Import dialog and browse for the file), and Persephone will auto-detect its file type: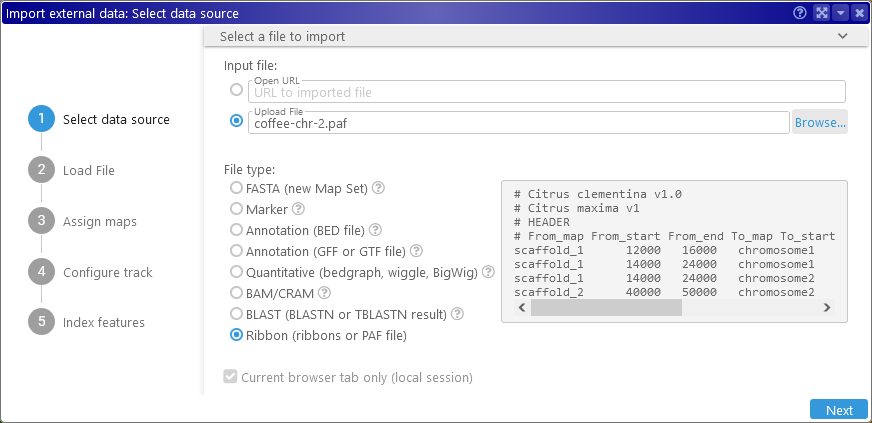
On the following screens, you will be prompted to assign the correct map sets to both the query and the target sequences in the imported file, in order to match the map names in the file with the correct maps in each map set. Most of the time, this process will happen automatically; however, minimap2 may not always handle spaces in FASTA headers correctly. Thus you may need to rename the maps in your input file, then match some of the maps by hand: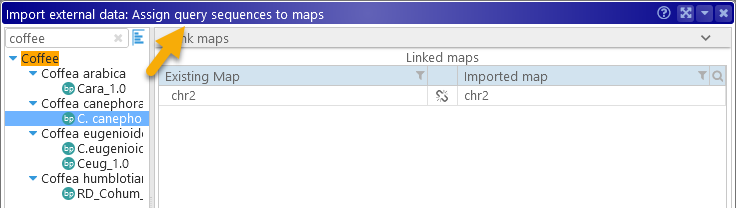
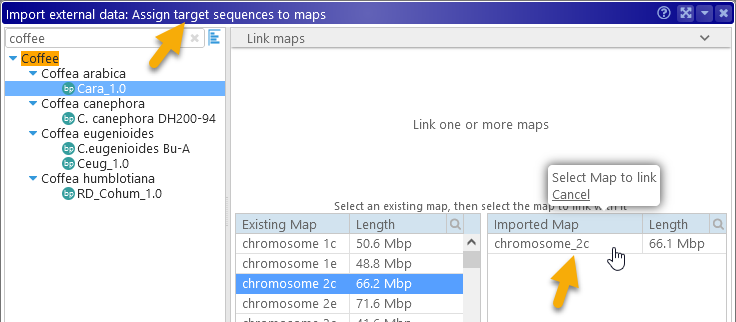
Finally, the imported alignments will be displayed as a Synteny track: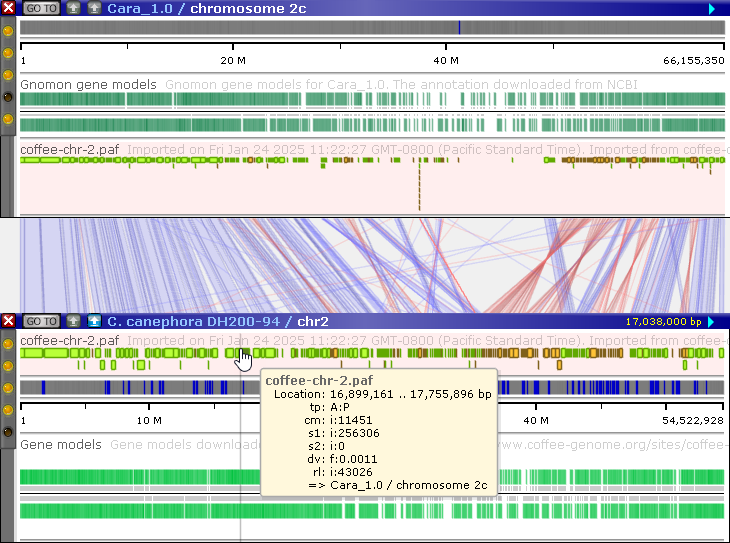
If needed, you can generate a more detailed alignment using Instant BLASTN. For example, you can zoom in on the repeat around 37 Mbp, then press the Instant BLASTN button on the bottom map to generate a nucleotide-level alignment: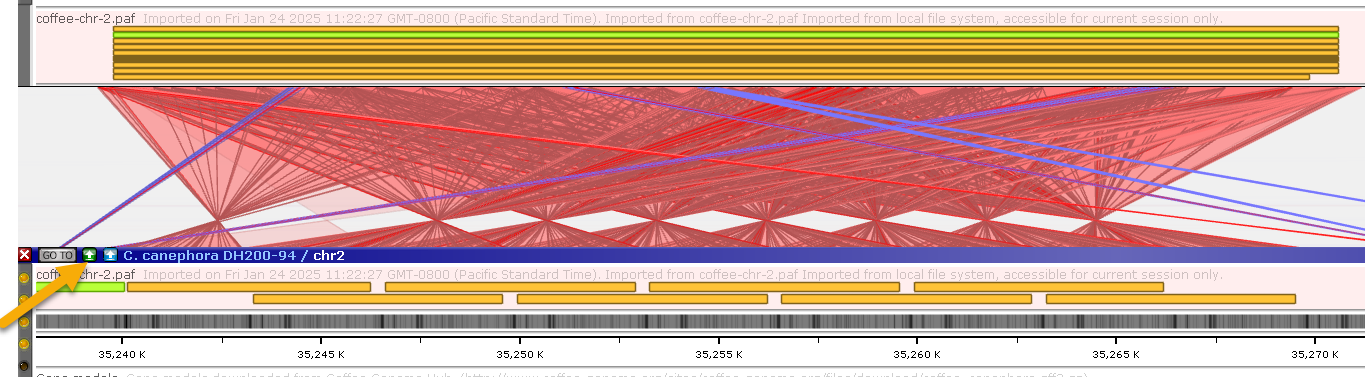
Loading a Ribbon file
Persephone's ribbon file format is a tab-separated CSV file with an optional header. The file describes ribbons between maps belonging to a pair of map sets:
# C. canephora DH200-94
# Cara_1.0
# HEADER
#FROM_MAP FROM_START FROM_END TO_MAP TO_START TO_END RIBBON_NAME Color
chr2 21319760 21393620 chromosome 2c 19259583 19326412 A Red
chr2 21407044 21462494 chromosome 2c 19340426 19392305 B Green
chr2 21483190 21497662 chromosome 2c 19438579 19452965 C #99ccff
If the header is present, it must contain exactly 4 lines:
1. The exact name or accession of the top map set.
2. The exact name or accession of the bottom map set.
3. The literal string "#HEADER".
4. The list of columns in the file; this is for documentation purposes only, and has no effect on parsing the file.
The body of the file describes the ribbons; one ribbon per line, specified by 6 to 8 tab-separated columns:
1. The exact name or accession of a map belonging to the top map set.
2. The starting position of the ribbon on this map (in bp, 1-based).
3. The ending position of the ribbon on this map (in bp, 1-based).
4. The exact name or accession of a map belonging to the bottom map set.
5. The starting position of the ribbon on that map (in bp, 1-based).
6. The ending position of the ribbon on that map (in bp, 1-based).
7. (optional) The name of the ribbon (to be displayed in the popup balloon for this ribbon's bar on the Synteny track).
8. (optional) The ribbon's color. This can be a color name such as "Red" or an HTML color code such as "#99ccff".
To import this file, drag-and-drop it onto Persephone's browser tab, or browse for it in the Import dialog. If the file contains a valid header, and all of the map set names and map names in the file match those in Persephone's database (or user storage), then you could click Next and then Finish now to import the file immediately: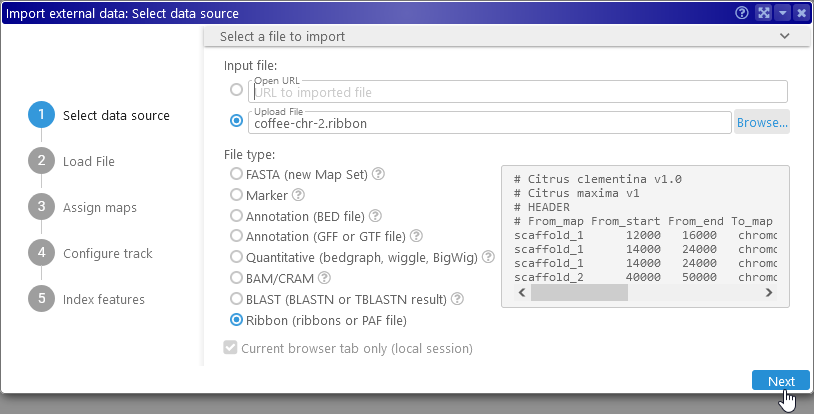
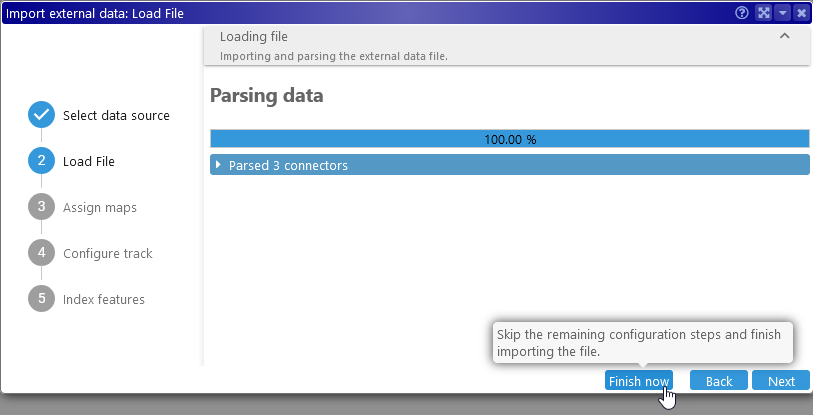
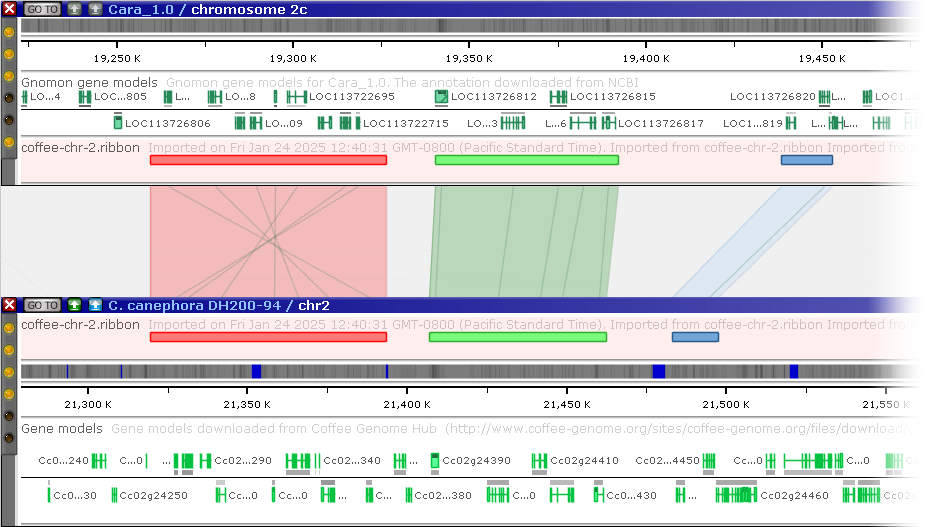
The sample file above will produce three ribbons:
Note
You can also load Ribbon files into Persephone's database using PersephoneShell.
