Web Persephone: Import
You can load a variety of data files directly into Persephone, either by drag-and-dropping them onto the browser window or by manually opening the Import dialog. Unlike data that is loaded into the database via PersephoneShell, these files will not be shared with other users, but will be accessible only by you. Persephone provides two different ways of storing the imported files:
- Long-term private storage: compressed copies of imported files are stored in your private user storage space on the server. The loaded data will be available at all times (until you choose to delete it), and will persist across different browser sessions.
Note
If you are using the public instance of Persephone (https://web.persephonesoft.com), you will need to create a free user account before you can load data into private storage.
Depending on your Persephone configuration, your user account may have limited storage quota for imported files. You can review all of your imported data in the User data dialog (where you can also delete any files that are no longer needed).
- Local browser session: the imported files are temporarily loaded into your current browser tab; when you close the tab, the loaded files will vanish (although you can always re-import them again next time). This option is best suitable for previewing large BAM files, since data loaded in this way does not count against your storage quota.
In most cases, imported files will be displayed as tracks on existing maps (alongside the tracks already available in the database); however, you can also create a new map set by loading sequences from a FASTA file.
To import a local file, you can drag-and-drop it directly onto Persephone's browser tab:

Alternatively, you can manually open the Import dialog from the main toolbar:

You can also construct URLs to link directly to Persephone with a pre-selected input file, as described here.
1: Select the input file
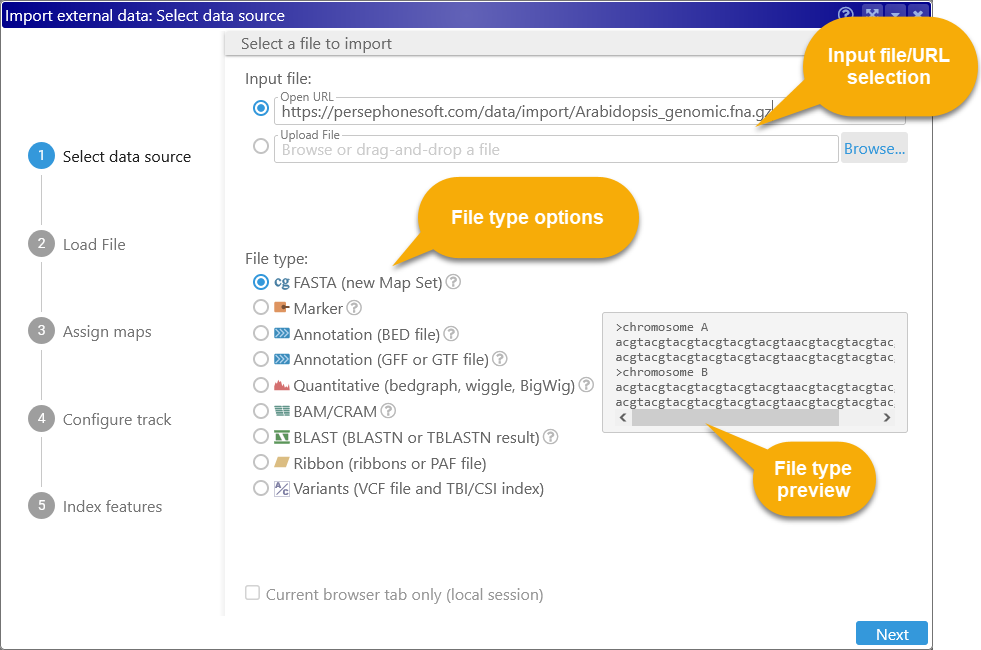
In this dialog, you can paste in a URL into the Open URL box, or click the Browse button to browse for a local file (if you drag-and-dropped a file, it will be filled in automatically).
Note
Only publicly accessible URLs can be imported into Persephone.
Persephone supports loading raw data files as well as files compressed by most popular compression tools, such as GZip (.gz), ZIP (.zip), or BZ2 (.bz2). It will attempt to select the appropriate file type based on the file extension; alternatively, you can manually select the appropriate file type. The file type preview box will display a small sample file of the selected type. The specific file types supported by Persephone are discussed in more detail below.
Check the Current browser tab only checkbox to load the file as temporary data, accessible only in the current browser tab. This option is not available for some file formats.
2: Load and parse the file
Click the Next button to begin loading the file. Some file formats will display additional options here (as described below); but in most cases you will see a dialog similar to this one:

Click the  button to show verbose log messages describing the process of loading and parsing the file; you can also do this when the process is complete:
button to show verbose log messages describing the process of loading and parsing the file; you can also do this when the process is complete:

This log will also show detailed error messages if the loading process fails.
3: Assign maps
Click Next again to describe how maps in the imported file should be treated. For most file formats (with the exception of FASTA), this means selecting a map set, and assigning map names from the imported file to maps in the map set. The initial view displays all maps in the input file, along with their length and projected size on disk (as always, this table supports all standard search and filtering controls).
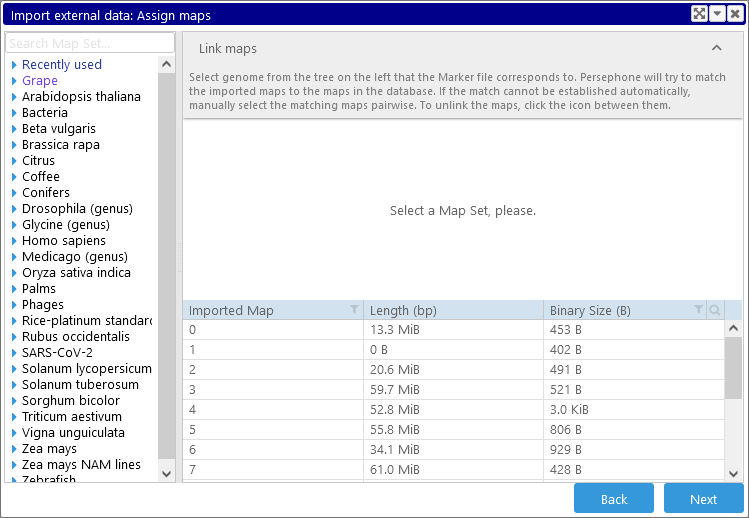
First, select a map set in the map set tree on the left-hand side; you can use the quick-search bar to speed up the process:
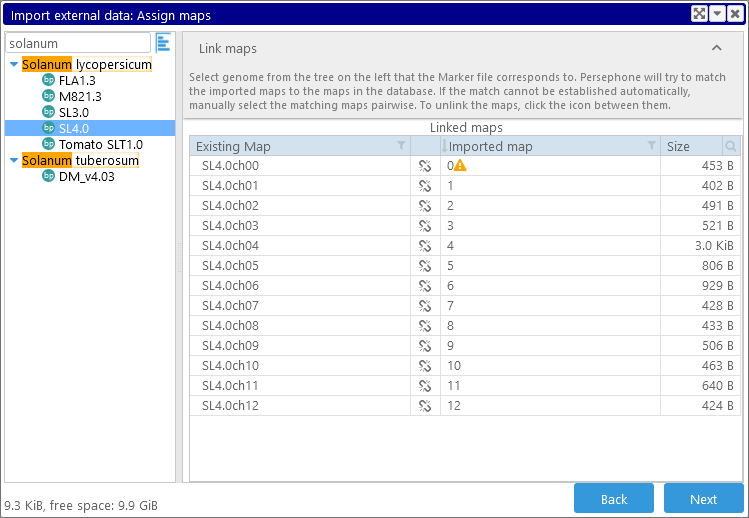
In this case, Persephone was able to automatically find matches for every map in the input file. However, note that map "0" has a  warning icon next to it. Mouse over the icon to display a reason for the warning:
warning icon next to it. Mouse over the icon to display a reason for the warning:

This means that a feature in the imported map (e.g. a marker, an annotation, a quantitative block, etc.) lies outside of the boundaries of the corresponding map in the database. This usually happens when the wrong version of an organism is selected. In this example, selecting SL3.0 instead of SL4.0 fixes the issue:

In this case, Persephone was able to automatically find matches for all of the maps in the input file. However, this isn't always possible. For example, Persephone failed to automatically find matches between all of the maps maps in the Wheat IWGSC v2.1 map set (Triticum aestivum), and maps in a VCF file:
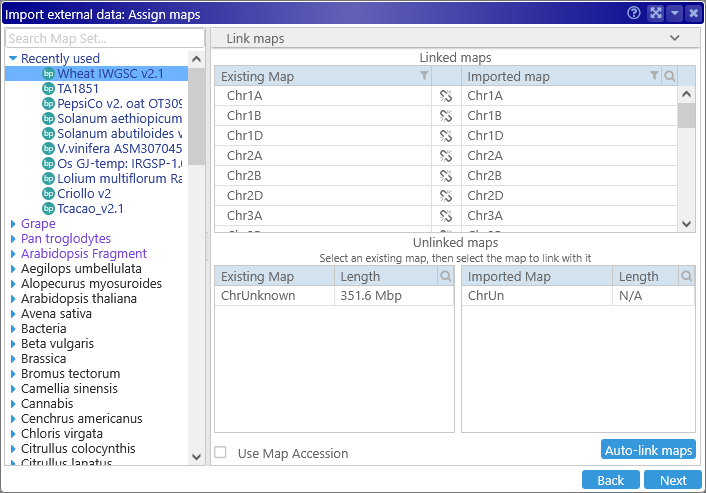
In such cases you can link the maps manually. Select an existing map in the database (in the table on the left), then select the matching map from the input file (in the table on the right). The two maps will be linked:
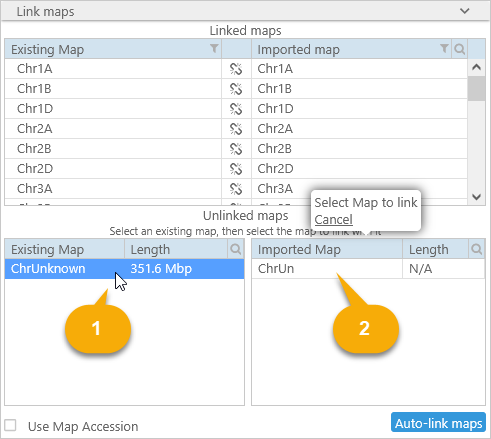
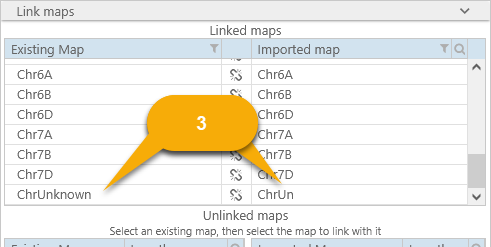
If you made a mistake, click the  button to un-link the maps. You should link at least one map to continue loading the file; however, note that only linked maps will be imported; any other maps will be discarded.
button to un-link the maps. You should link at least one map to continue loading the file; however, note that only linked maps will be imported; any other maps will be discarded.
You can also check the Use Map Accession checkbox to match the maps based on their accession numbers instead of human-readable names. This can be useful when loading data from sources such as NCBI, who typically publish their data files using accession numbers rather than names.
4: Configure track and select indexing options
Click the Next button to configure track options, including the track name, color, and description (you can always re-visit these options later in the Configure Track dialog). Depending on the file format, you can also choose whether to index the imported data for Search (if the file contains named features such as markers) or BLAST (if the file contains genomic sequences or gene annotations). For example, the GFF format supports both indexes:
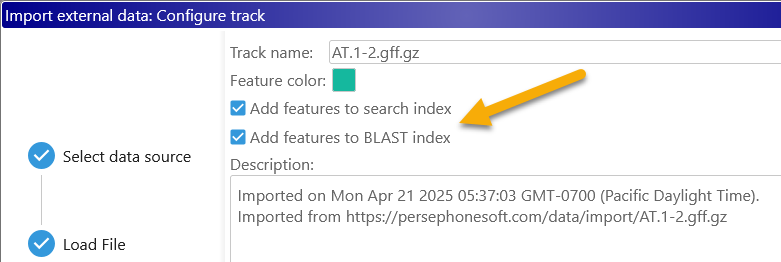
Indexing is always enabled by default, but you may choose to skip it to save time. For example, when loading a track containing repeats,
5: Index features and Finish
If you have chosen to create any of the indexes, click Next to index your newly imported data; the indexing process can take several minutes. Finally, you can choose to automatically open one of the imported maps:

The imported track will be displayed on the map. All imported tracks are highlighted with a light teal background if they were loaded into long-term private storage:

...and with a light pink background if they were loaded into the local browser session:

Supported file formats
Persephone currently supports importing the following file formats:
- FASTA: Maps with genomic sequences; creates a new map set.
- Marker: Tab-delimited file containing named intervals on maps.
- Annotation: Gene annotations (or transcripts) on physical maps, in GFF/GTF or BED formats.
- Quantitative: Regions with numeric values on maps, in BedGraph or Wiggle/BigWig formats.
- BAM: Multiple reads aligned to physical maps, usually containing mutations, in BAM/CRAM formats.
- BLAST: Results of running BLASTN or TBLASTN (BLASTP and BLASTX are currently not supported)
- Ribbon: Syntenic blocks between maps (such as alignments generated by minimap2), in PAF or Persephone's Ribbon formats
- VCF: Variants (e.g. SNPs and Insertions/Deletions) aligned to physical maps.
