Web Persephone: Tips for using BLAST parameters
Quite often the default BLAST parameters do not give an optimal result. For example, High Scoring Pairs (HSP) can accidentally merge the matches for neighboring exons:
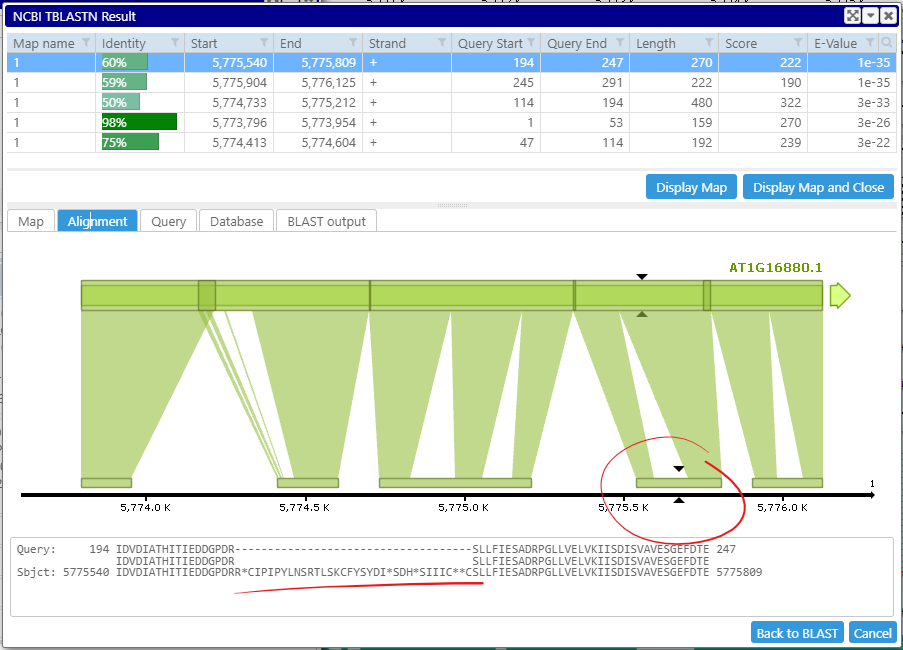
Cross-species protein matches (TBLASTN) emphasize conservation of the coding sequences. Having correct boundaries of the HSPs can be crucial for accuracy of gene prediction where an HSP is expected to correspond to an exon.
To split the fused HSPs, we need to increase the penalty for opening and extending the gaps by adding these parameters:
-gapopen 11 -gapextend 2
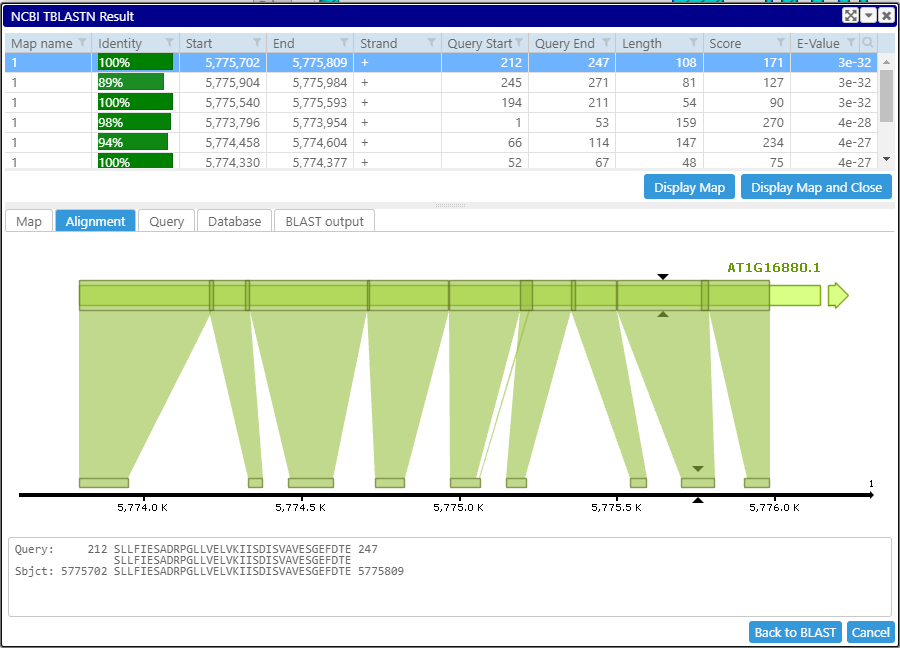
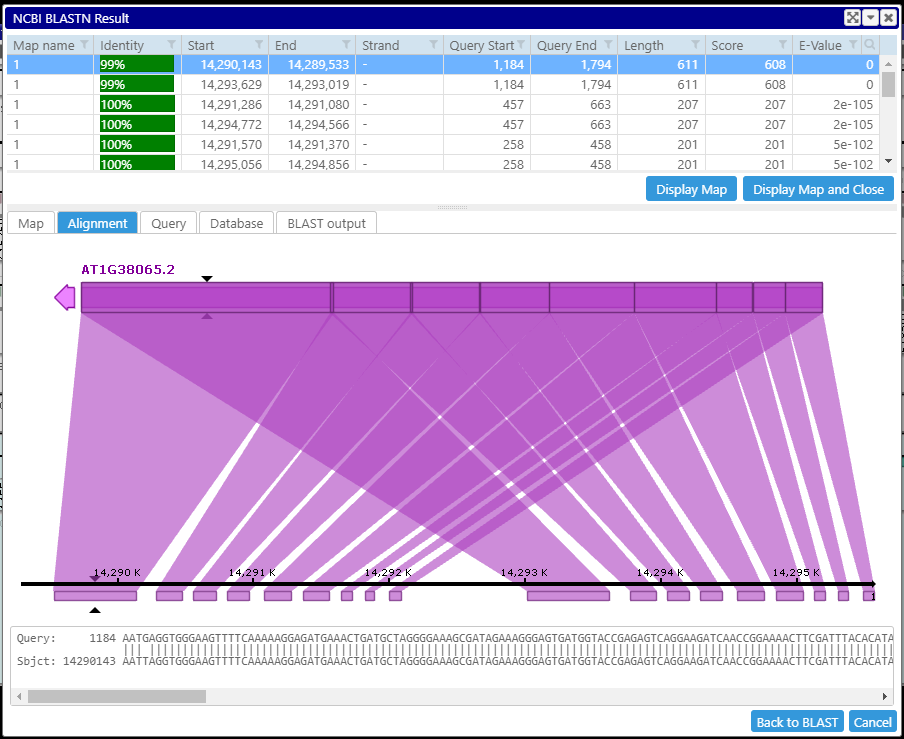
Another, frequently overlooked, parameter is culling_limit. It specifies how many times the same query sequence can "participate" in an HSP. Having culling_limit greater than one will help identify sequence repeats.
culling_limit=2:
culling_limit=1:
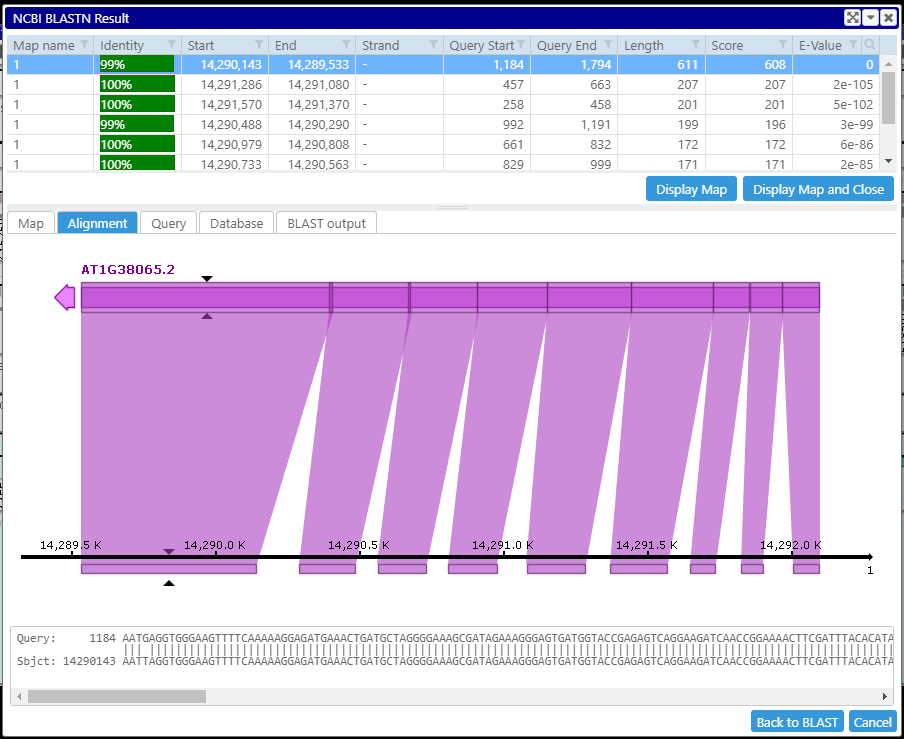
Setting culling_limit to 1 helps with cleaning up the results of Instant BLAST.
A proper set of BLASTN parameters should be used when searching for primer targets.
