Add Functional Annotation
If the newly loaded gene models lack functional annotation, you can run the command create function. For each gene, it will try to find matches to well-characterized Swiss-Prot entries, which are manually annotated and reviewed by experts, ensuring high-quality and reliable information about proteins. This way, the functional annotation will be recorded as the new qualifier SwissProt match.
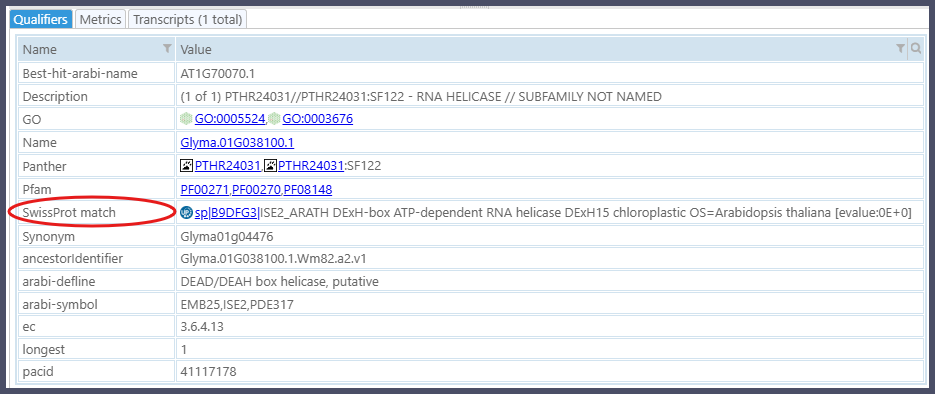
In our use case, the command to find matches is:
PS> create function "/Glycine max/Wm82.a4.v1"
Assigning functional annotation by match to well-characterized proteins from SwissProt
- SwissProt file is present: passed
- Track Gene models
/data/OpenPlants/NCBIBlast/bin/blastdbcmd -db "/data/OpenPlants/BlastDB/34_Phytozome13_P" -entry all -out "/HDD4Gb/Temp/_blast/34_Phytozome13_P"...
┌───────────────────────────────────────────────────────────────────────────────────────────────┐
│ │
│ SwissProt. Downloaded from The UniProt Consortium │
│ https://ftp.uniprot.org/pub/databases/uniprot/knowledgebase/complete/uniprot_sprot.fasta.gz │
│ UniProt: the Universal Protein Knowledgebase in 2023 │
│ Nucleic Acids Res. 51:D523–D531 (2023) │
│ https://uniprot.org │
│ │
└───────────────────────────────────────────────────────────────────────────────────────────────┘
Running DIAMOND...
First lines of the output:
ANNOT_ID DESCRIPTION
-------------------------------------------------------------------------------------------------------------------------------------------------------------------
2843853 Protein PHYLLO, chloroplastic OS=Arabidopsis thaliana [evalue:2.82E-15]
2843855 Photosystem II stability/assembly factor HCF136, chloroplastic OS=Oryza sativa subsp. japonica [evalue:1.86E-16]
2843859 Protein PHYLLO, chloroplastic OS=Arabidopsis thaliana [evalue:8.63E-26]
2843861 Protein FAR-RED IMPAIRED RESPONSE 1 OS=Arabidopsis thaliana [evalue:1.16E-102]
2843862 Protein FAR-RED ELONGATED HYPOCOTYL 3 OS=Arabidopsis thaliana [evalue:7.39E-142]
2843866 Chloroplast envelope membrane protein OS=Rhodomonas salina [evalue:6.16E-37]
2843871 TORTIFOLIA1-like protein 2 OS=Arabidopsis thaliana [evalue:1.55E-179]
2843874 Cycloartenol synthase OS=Oryza sativa subsp. japonica [evalue:0E+0]
Display [N]ext 15 lines; [S]ave functional annotation; [ESC] - cancel: S
Saving qualifiers (SwissProt match) to the database...
Stored 65,537 records
Do you want to nominate SwissProt match qualifier as gene function? (Y/N) Y
DATA_VERSION updated
Success
