Web Persephone: Annotation export
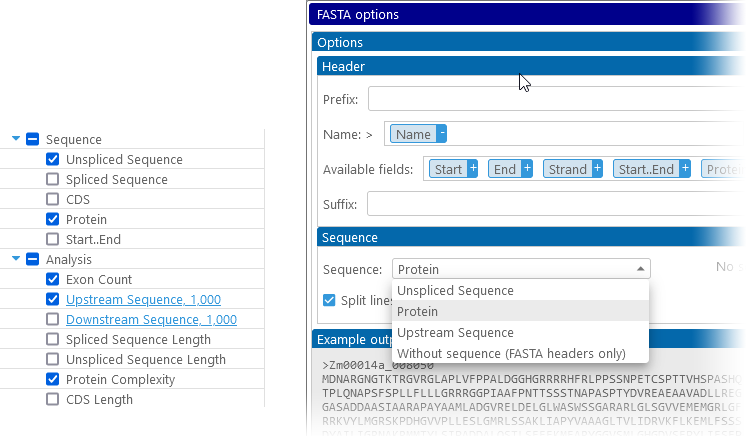
In addition to the common export options, Annotations provide the following sets of columns for export:
- Basic properties: includes all of the properties of the annotation, including its name, location on the map, and CDS coordinates (if any).
- Name: this is the preferred name of the specific transcript. Additional names may be found in Qualifiers. If no names have been loaded for this annotation, its numeric ID will be used instead.
- Group Name: the name of the entire gene (which may be shared among multiple transcripts). Some annotations may not have group names (though they will always have individual names or IDs).
- Sequence: lists all of the available sequences for the annotation. Note that although these sequences may be truncated in the preview table, they will always be exported in full. Note that CDS and Protein sequences may be unavailable if the annotation lacks CDS.
- Start..End: exports annotation coordinates (in bp) as a single value, e.g. "2759..7959". This value can be pasted directly into e.g. the Go To dialog.
- Analysis: provides various types of analysis that are not directly encoded in the annotation metadata.
- Exon Count: The number of exons for this annotation.
- Upstream/Downstream sequence: Click these columns to set the length of the upstream or downstream sequence to export:
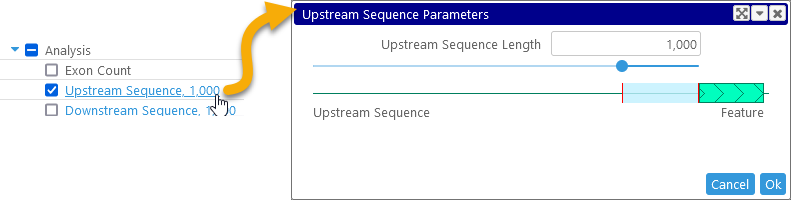
You can set the desired sequence length by moving the slider, or by entering the value directly into the textbox; at most 5,000 upstream/downstream bp can be exported. - Protein Complexity: complexity of the protein based on entropy of 2-mers (as a floating point value).
- Unspliced Sequence Length / Spliced Sequence Length / CDS length: the length of the corresponding sequence, in nucleotides.
Output file formats
Annotations can be exported as CSV, FASTA, GENBANK, or GFF. Note that in order to provide the sequence for FASTA, you must first select it in the column chooser: