Web Persephone: The Annotation Details dialog
This dialog, also known as the "Gene Card", displays all known properties of an annotation, including its qualifiers, sequences, metrics, isoforms, and orthologs (if any). To open the dialog, click an annotation feature on the Annotation track (or anywhere else in Persephone, e.g. in the Search results or Export tables). If the annotation's track is visible, the annotation you clicked will be outlined with a solid border:
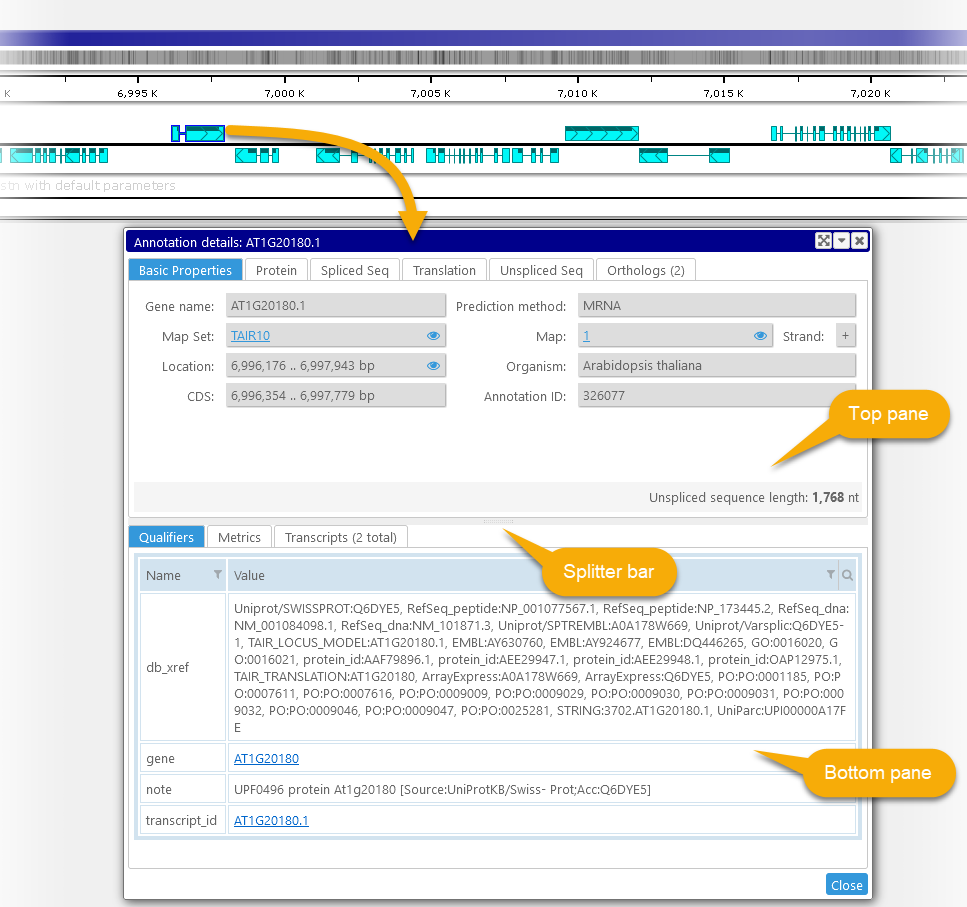
The Annotation Details dialog is split into two halves: the top pane, and the bottom pane; you can drag the splitter bar with the mouse to resize them (as shown above). Both panes contain several tabs. By default, the Basic Properties tab will be selected on the top, and the Qualifiers tab will be selected on the bottom; however, the dialog will "remember" which tabs were selected the last time it was opened, and select the same tabs the next time it is opened.
Top: Basic Properties
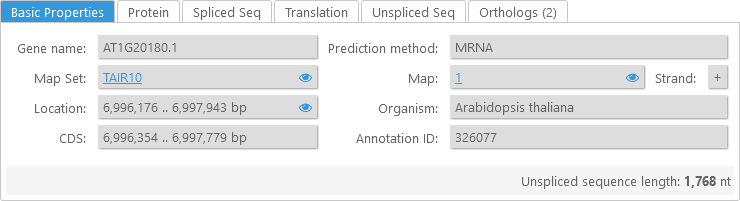
This tab displays all the basic metadata that is known about the annotation.
|
Gene Name: |
The specific name of the currently selected transcript; note that there could be multiple transcripts for a gene. |
The method that was used to predict this annotation; typically, it will be the same as the name of the annotation's track. |
|
|
Map set: |
The map set where the annotation belongs. Click the map set name to open its Map set details dialog; click the |
Map: |
The map where the annotation belongs. Click the map name to open the Map details dialog; click the |
|
The start and end coordinates of the annotation, in bp. Click the |
Organism: |
This annotation's organism, as defined in its map set. |
|
|
CDS: |
If the annotation has a CDS, shows its start and end coordinates, in bp. |
|
|
|
|
Unspliced sequence length: |
The length of this annotation's unspliced sequence, in nucleotides. |
|
Gene quality marks
If the annotation track is configured to display gene quality marks, the quality of each annotation will be prominently displayed among its properties:
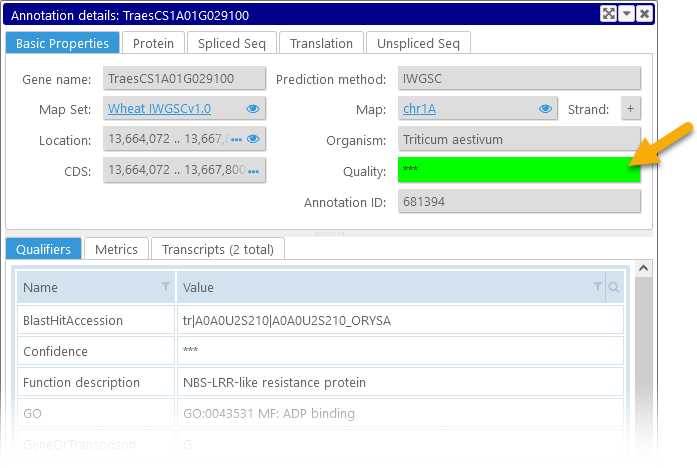
The color of the quality box depends on the value of a specific qualifier (or a set of qualifiers), as configured by the Persephone administrator. The value of the relevant qualifier is shown inside the box (in this case, "***"). You can view the full list of this annotation's qualifiers on the Qualifiers tab in the bottom portion of the dialog.
Top: Protein
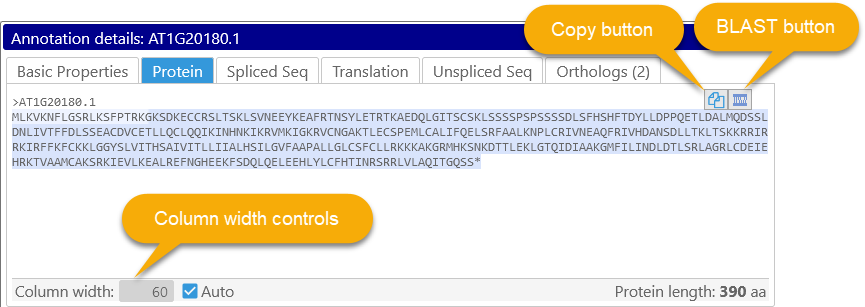
If the annotation has CDS (as most gene models do), then you can see its auto-generated protein translation here (in FASTA format). The sequence is highlighted to emphasize individual exons, with alternating bands of lighter and darker color. You can double-click anywhere inside the sequence for an individual exon to select the entire exon:
You can also press Ctrl-A (Command-A on Mac) to select the entire sequence. Text selection is synchronized across all of the sequence tabs in this dialog, as well as the Metrics view.
Click the copy button (![]() ) to copy the entire protein sequence to the clipboard; click the BLAST button (
) to copy the entire protein sequence to the clipboard; click the BLAST button ( ) to open the BLAST form and pre-fill it with the current protein sequence for TBLASTN.
) to open the BLAST form and pre-fill it with the current protein sequence for TBLASTN.
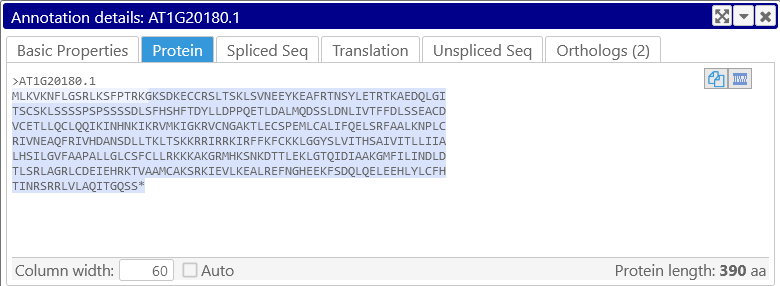
You can also change the number of sequence characters shown per line; if the Auto checkbox is checked, the sequence will automatically wrap to take up the entire available space. Uncheck the checkbox to set the column width manually, e.g. at 60 characters:
Top: Spliced Seq
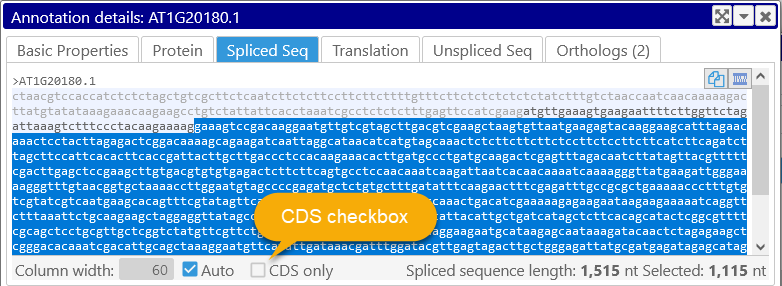
This tab shows the spliced DNA sequence of the annotation; individual exons and are highlighted in bands of alternating lighter and darker color. By default, the UTRs are included in the sequence and highlighted in light gray color; you can double-click a UTR to select it (similarly to selecting an exon). You can also hide the UTRs by checking the CDS only checkbox.
Click the copy button (![]() ) to copy the entire sequence to the clipboard; click the BLAST button (
) to copy the entire sequence to the clipboard; click the BLAST button ( ) to open the BLAST form and pre-fill it with the current DNA sequence for BLASTN.
) to open the BLAST form and pre-fill it with the current DNA sequence for BLASTN.
Top: Translation
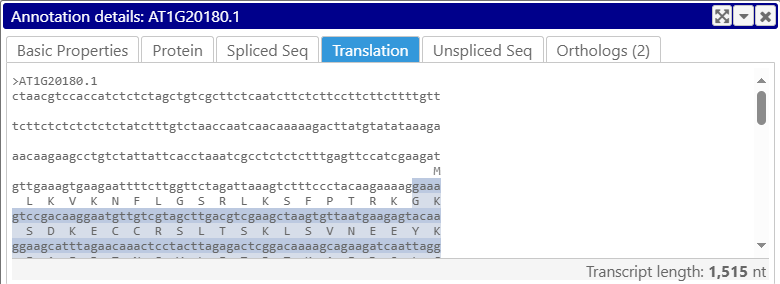
If the annotation has CDS, this tab will show its spliced sequence alongside its protein translation; in this view, the sequence is automatically wrapped at 60 characters. You cannot directly select the in this tab, but any previous selection will be preserved; as usual, you can also use the Metrics view to select the sequence (or switch to another sequence tab and select the sequence there).
Top: Unspliced Seq
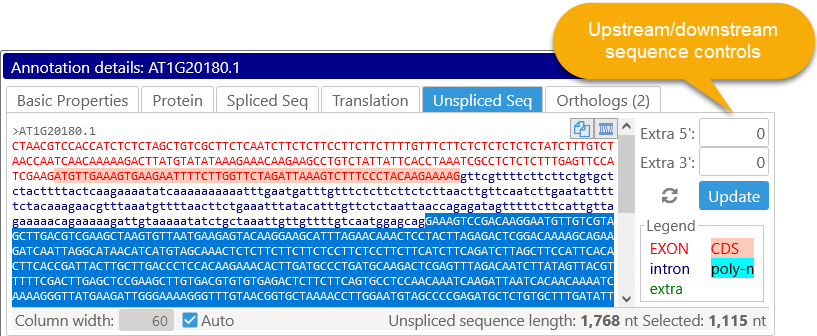
This tab displays the unspliced sequence of the annotation, color-coded based on sequence fragment type (as shown in the Legend on the bottom-right). You can also select an additional length of sequence at the annotation's 5' and/or 3' ends, e.g. to display a potential promoter. This sequence will be shown in green:
Click the  button to reset both fields to zero.
button to reset both fields to zero.
You can also copy, BLAST, and format the sequence (in the same way as described above for the Protein and Spliced Seq tabs). As usual, you can double-click the individual sequence fragments (exons, introns, UTRs, and upstream/downstream sequences) to select them.
Top: Orthologs
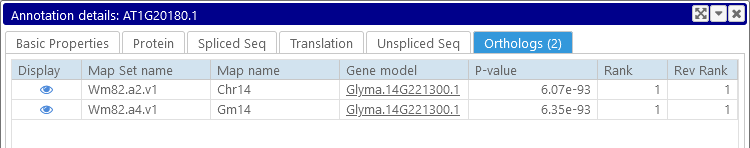
If the annotation has any orthologs, they will be shown here (otherwise, the Orthologs tab will be hidden). Every row in this table displays an ortholog, along with its metadata. Click the  icon to open the map with the orthologous annotation; highlight the ortholog; and zoom into its location:
icon to open the map with the orthologous annotation; highlight the ortholog; and zoom into its location:
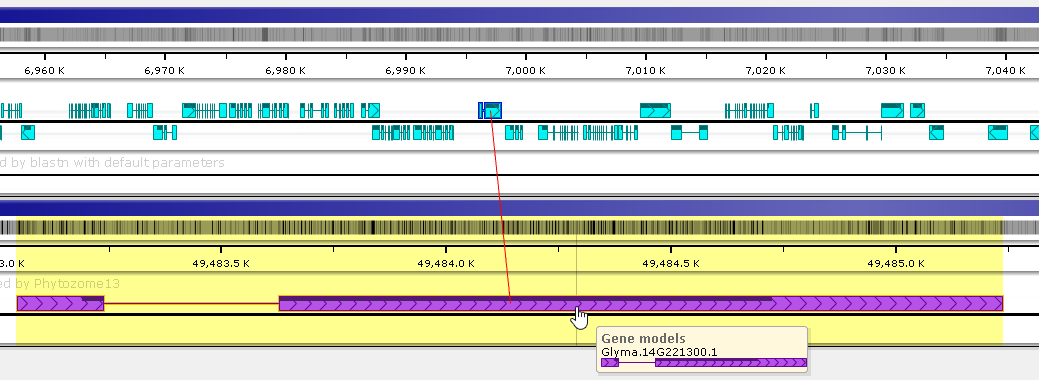
The ortholog will be automatically connected to the current annotation by a line connector.
Click the name of the ortholog to open its own Annotation details dialog (alongside the current dialog).
Bottom: Qualifiers
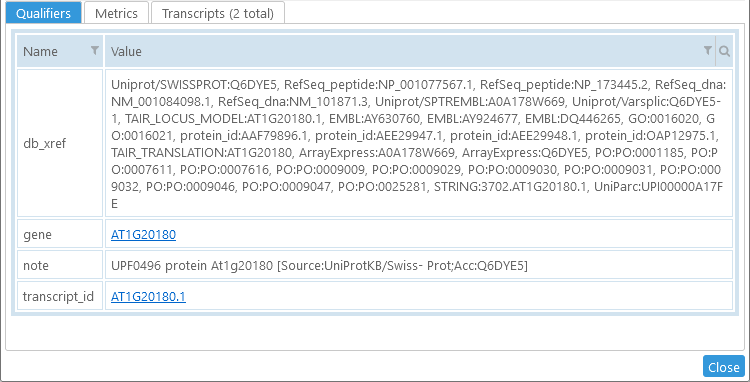
If the annotation has any qualifiers, they will be displayed here. Qualifiers are simple key/value pairs that can represent all kinds of metadata, such as alternative names for this annotation; its gene function; database cross-references; and researcher's notes. Some of the qualifiers may be displayed as links; these links typically lead to an external database from where the gene was loaded into Persephone. In this example, the gene is linked to TAIR; however, these qualifier links can be configured individually for each map set in PersephoneShell.
Note
The table of qualifiers is equipped with the standard table sorting and filtering controls.
Bottom: Metrics
Displays the Metrics view for this transcript, to assist with measuring various sub-sequences of the annotation.
Bottom: Transcripts
Displays the Transcripts View for the entire gene, showing the current annotation alongside all of its other isoforms.
