Web Persephone: Connectors
When two or more maps are displayed side-by-side, they can often be connected to each other, e.g. by common features such as markers or orthologous gene pairs, syntenic regions between physical maps, homology ribbons, and other means.
Marker connectors
If the same markers are mapped to two different maps, Persephone will automatically draw line connectors between them (in this example, non-relevant tracks have been turned off for clarity, and the marker tracks are collapsed):
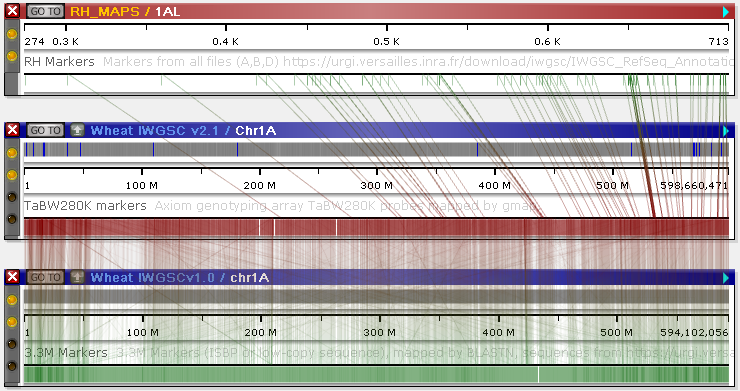
Note
Marker identity is handled slightly differently for markers loaded into the main Persephone database (usually via PersephoneShell), and markers that have been imported from user files (usually by drag-and-droping a file onto the browser window). These differences are described in more detail here.
You can change the opacity of line connectors in the Settings dialog; for example, this can be useful when connectors are so dense that they cover up other map features. Select Tools | Settings from the main toolbar, then open the Display tab in the Settings dialog, and move the Connector opacity slider:
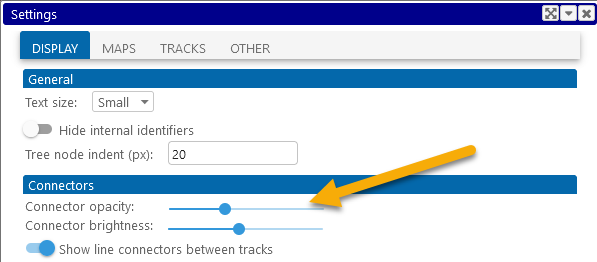
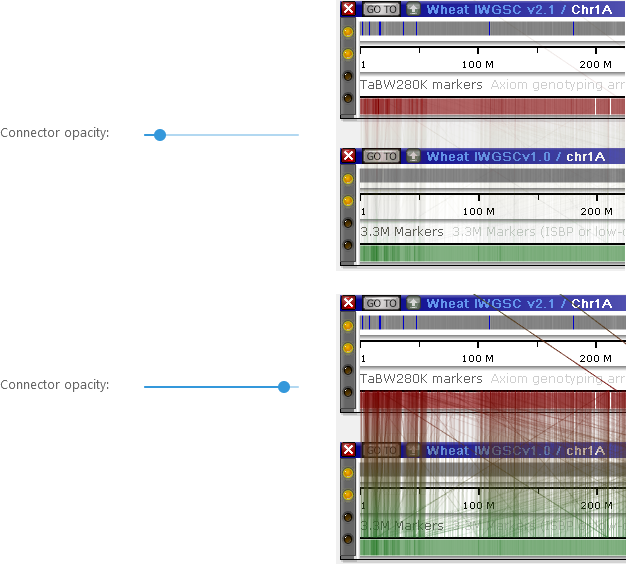
Moving the slider to the left makes connectors more transparent:
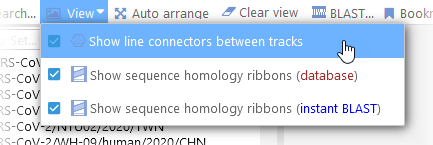
In the same tab of the Settings dialog, you can also hide line connectors altogether, or change their brightness (relative to the brightness of the markers they are connected to). You can also quickly toggle line connectors on and off in the View menu of the main toolbar:
If you mouse over a marker that has a line connector, the connector will be highlighted in red:

You can also right-click the marker, and select Align connected features from the context menu:
Doing so will simultaneously zoom and pan all of the maps to the location where the marker appears, in order to bring the marker into view:
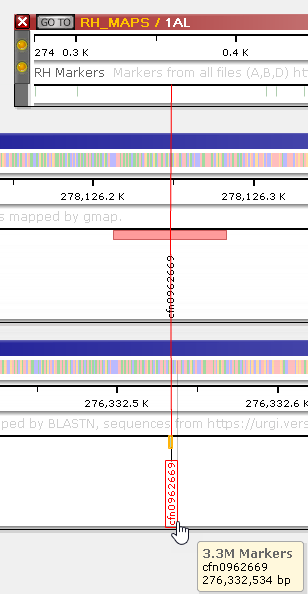
You can click the marker to open up the Marker details dialog, where you can find a list of all the maps where the marker appears.
Ortholog connectors
The line connectors linking orthologous annotations behave in a way similar to marker connectors:
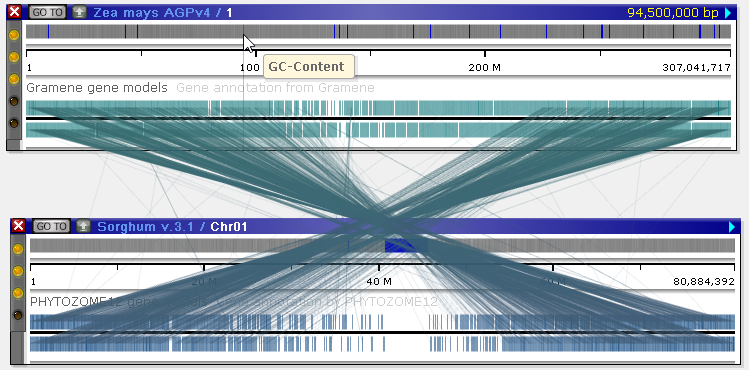
Note that, unlike marker connectors, ortholog connectors are separated by strand. As always, you can invert one of the maps to make connectors easier to see:
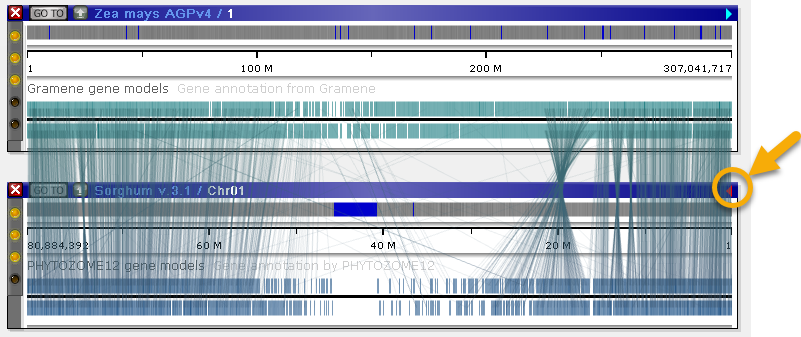
In all other respects, ortholog connectors behave the same way as marker connectors. You can view all known orthologs for any annotation by clicking the annotation, then navigating to the Orthologs tab of the Annotation details dialog.
Note
At present, there is no support for loading ortholog connectors for annotations that have been imported from user files (e.g. via drag-and-drop); however, you can still use the real-time BLAST tool to manually discover their orthologs.
Managing connectors
Whenever there are multiple connected tracks on the same pair of maps, Persephone will only connect the tracks that are closest to each other, hiding all connectors between other pairs of tracks in order to reduce clutter:

You can change this behavior in several ways.
Firstly, you can manually reorder tracks by dragging them in the Track Panel, to move the relevant tracks closer to the top or bottom edges of their maps:
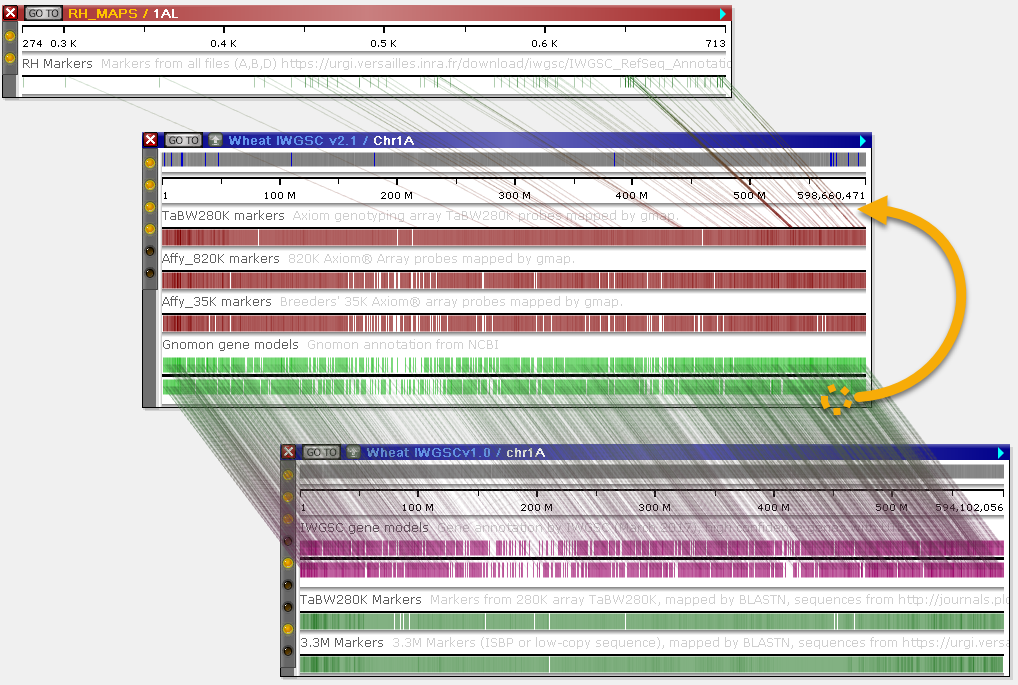
In this example, the marker track (TaBW280K markers) has been moved to the top of the map, and the annotation track (Gnomon gene models) to the bottom of the map; thus, Persephone prefers the connectors between the two annotation tracks (Gnomon gene models and IWGSC gene models), hiding the others.
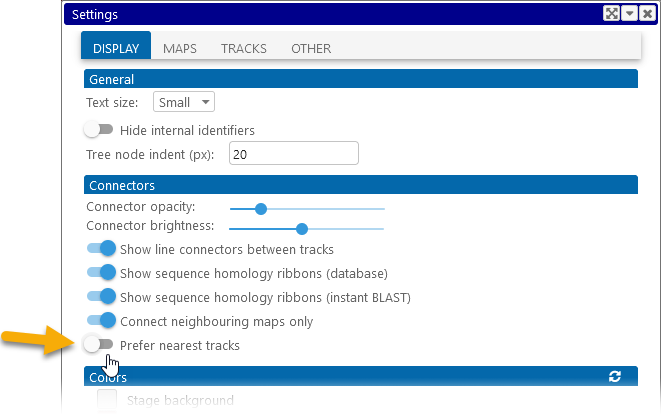
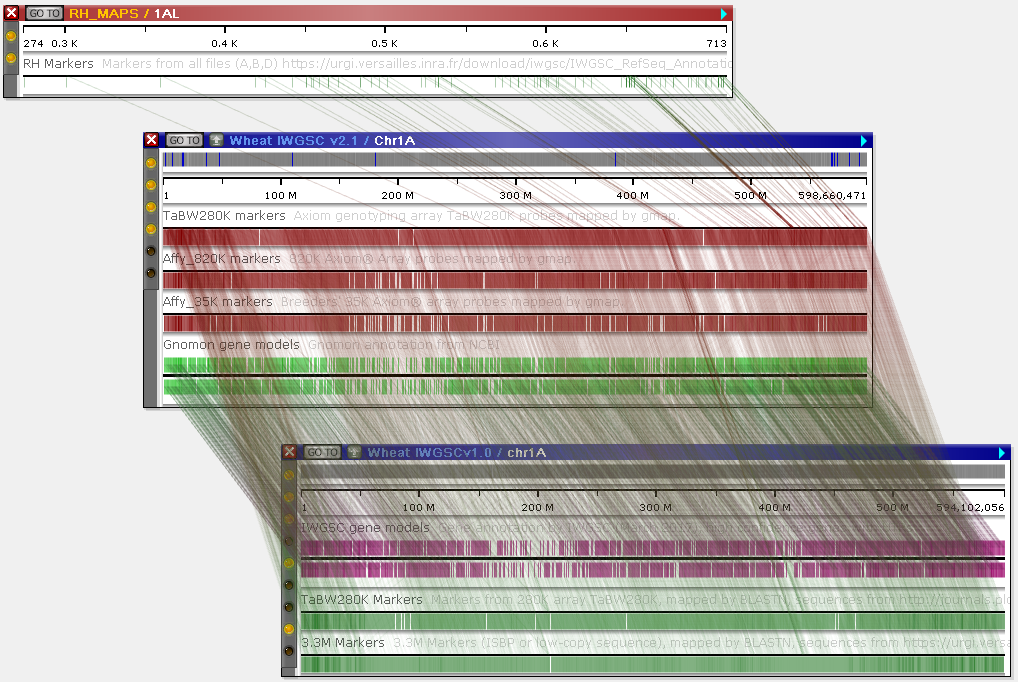
Secondly, you can reveal all possible connectors simultaneously. To do so, select Tools | Settings from the main toolbar to open the Settings dialog, navigate to the Display tab, and turn off the Prefer nearest tracks option:
Persephone will now show all available connectors between the two maps:
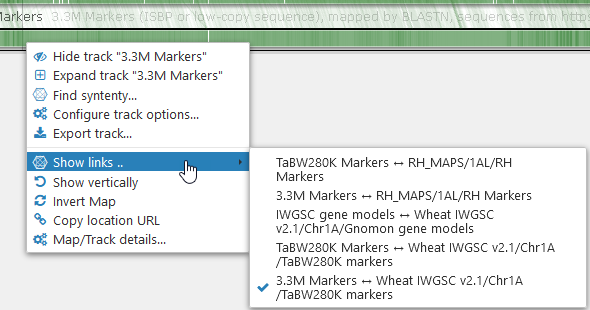
You can selectively turn specific connector sets on and off by right-clicking one of the connected maps, and selecting Show links from the context menu. You will see a list of all available connector sets for that map, with a checkbox next to each one; toggle the checkbox on or off to reveal or hide the corresponding connectors. For example, you could hide all connectors except the ones between the two marker tracks, 3.3M Markers and TaBW280K markers on the other map:

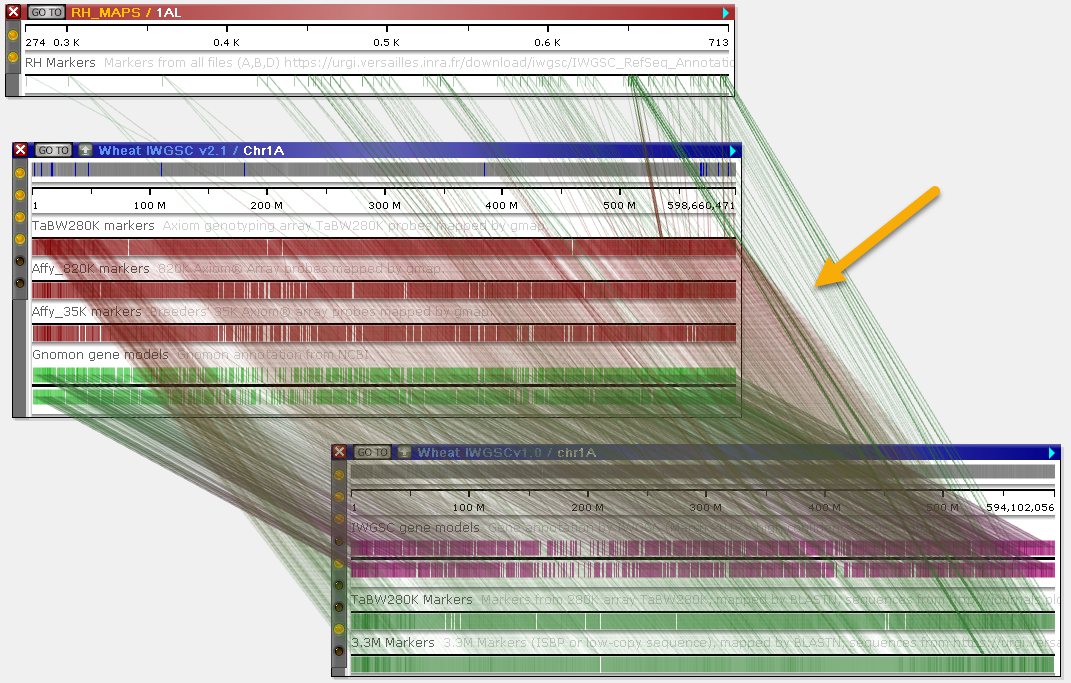
In addition, Persephone will normally only display connectors between neighboring maps (once again, in order to reduce visual clutter). For example, in this scenario there exist connectors between the genetic map on top (RH_MAPS / 1AL) and the physical map on the bottom (Wheat IWGSCv1.0 / chr1A); yet they are not shown, because the other map is "blocking" them. You can rearrange the maps (by dragging and zooming them) to reveal these connectors:
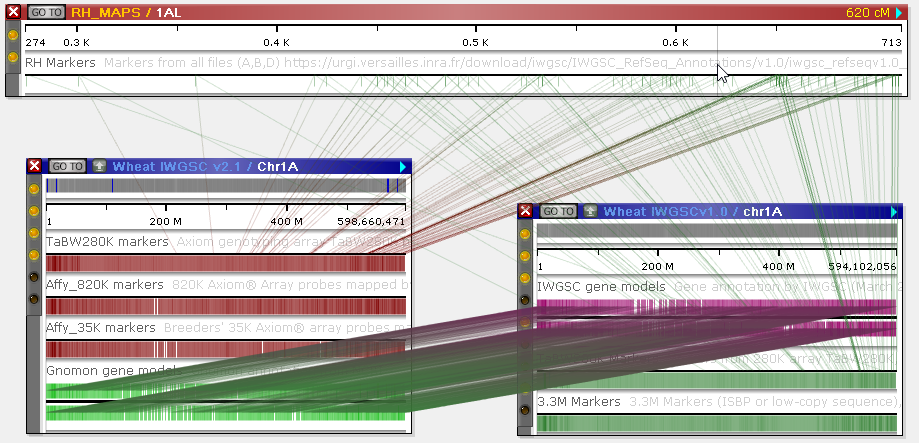
Alternatively, you can turn off the Connect neighboring maps only option on the Display tab of the Settings dialog:

Finally, you can use the MultiMap and Find Synteny tools to quickly navigate connected maps, as well as to find the best match for any map and/or track. These tools are described in more detail here.
To visualize all connectors between two map sets at a glance as a dot plot, use the Synteny Matrix tool.
Connector Appearance
Use the Settings menu to switch between curved and linear connectors and/or homology ribbons:

Note that curved connectors may render more slowly on older GPUs.

The images above also demonstrate the Draw connectors to map edges setting, which prevents connectors from overlapping with their connected maps -- at the expense of some additional screen real estate.
Homology ribbons
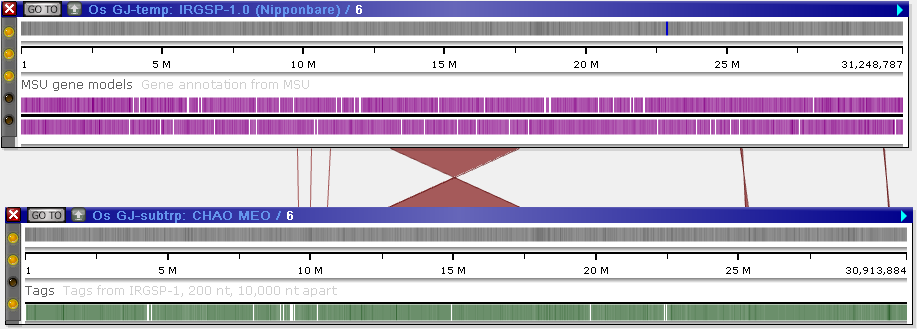
In addition to line connectors between tracks, some maps in the Persephone database can be connected by homology ribbons (these ribbons must be loaded in PersephoneShell). For example, such ribbons can indicate structural variations, such as inversions:
You can toggle these ribbons on and off from the View menu on the main toolbar, as well as in the Display tab of the Settings dialog:

This can be useful to reduce visual clutter while viewing regular marker or annotation connectors:
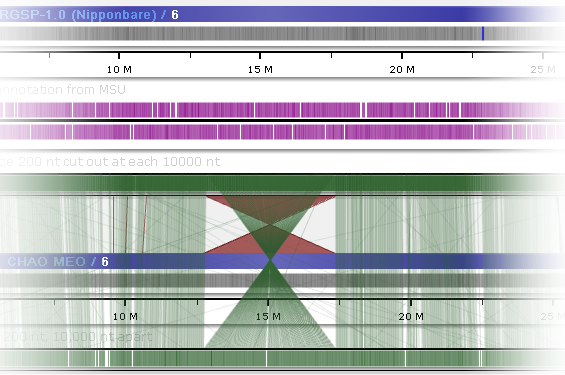
You can also change the color of these ribbons in the Settings dialog:
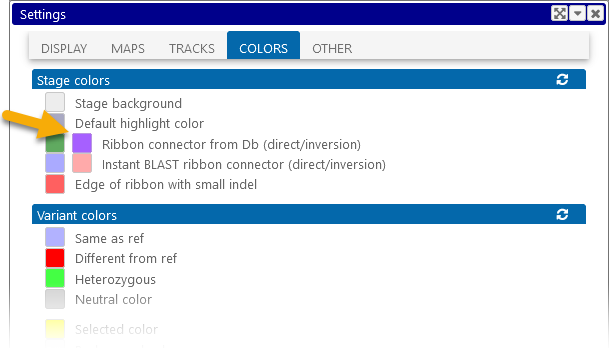
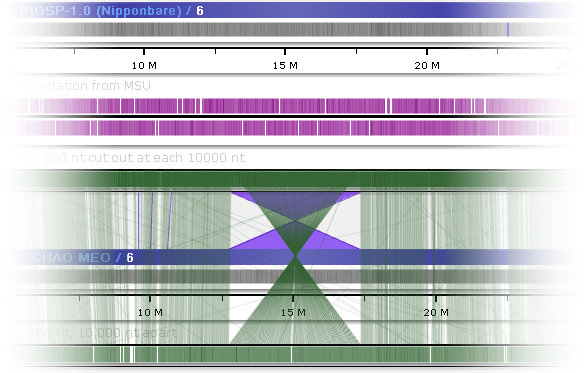
Click the  icon to return all colors to their defaults.
icon to return all colors to their defaults.
Instant BLAST and minimap2
You can generate similar ribbons on the fly by arranging two physical maps side-by-side, and aligning their sequences. You can align large sequence fragments (up to and including entire chromosomes) using minimap2, or drill down into smaller regions (about 1Mbp in length) and align them in greater detail using Instant BLASTN. These features are described in more detail here.
