Web Persephone: Virtual Tracks
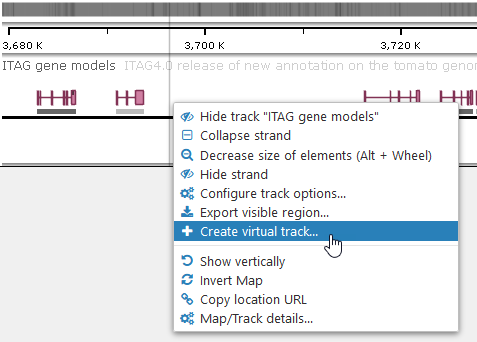
You can create Virtual Tracks by filtering and transforming features on existing tracks according to user-defined rules. To create a Virtual Track, right-click on a regular track, and select Create virtual track from the context menu to open the Virtual Track dialog:
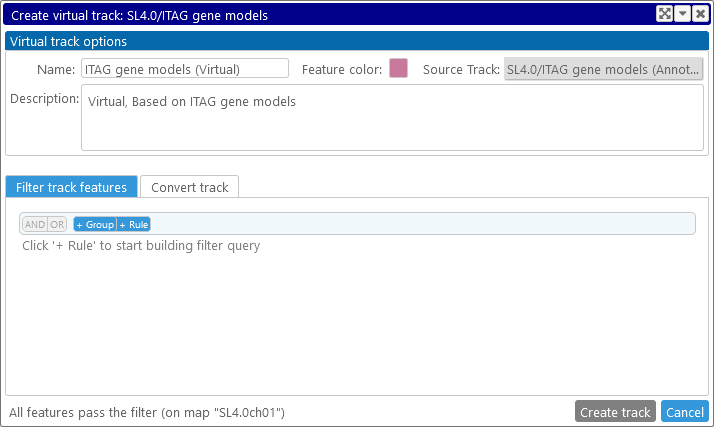
In this dialog, you can filter track features; convert them to another feature type; or do both at the same time.
Note
Virtual Tracks can currently only be created based on Annotation or Marker tracks.
Filtering track features (and other basic functionality)
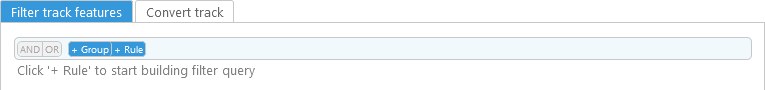
The two tabs in the bottom portion of the dialog contain filtering and conversion options; filtering (if any) is always applied first. To begin filtering track features, click the +Rule button:
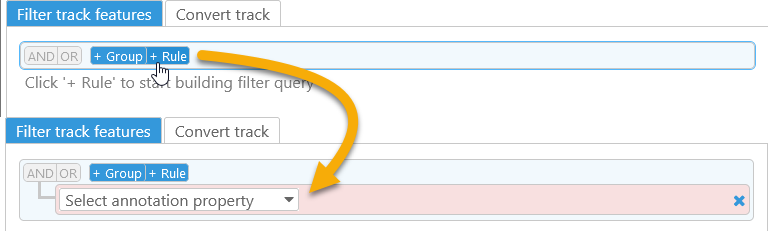
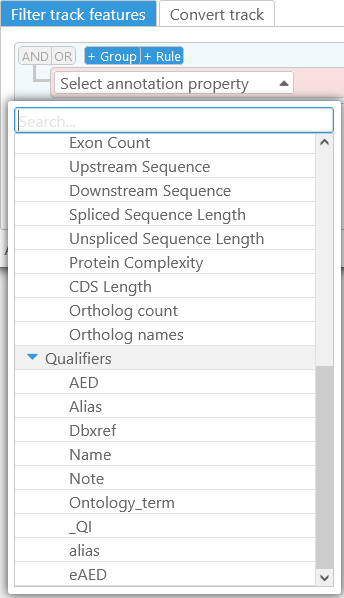
Then, select a property to filter by. In this example, the base track is an Annotation track, and thus you can base your filter on any property of gene annotations on this track, including basic properties (such as Start/End coordinates), sequences (e.g. the Protein sequence), or qualifiers. As usual, you can quick-search the list of properties by typing in the box at the top:
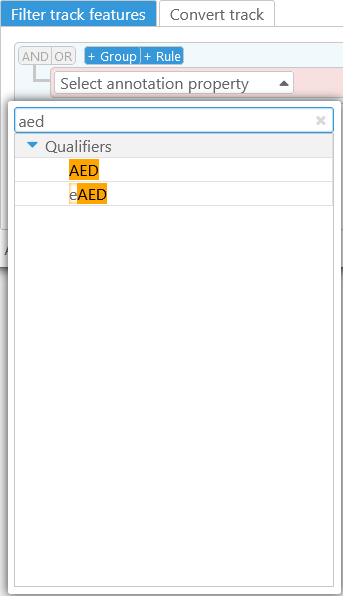
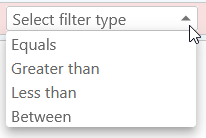
Select a property, such as the AED qualifier, and then select the appropriate Filter type. Because the AED qualifier is numeric, all of its available filter types are numeric comparison operators:
You can also click the  button to preview sample values for the chosen property (this button may not be available if there are too many different values for the chosen property). The table of sample values is sorted by frequency of occurrence (i.e. the number of times each value occurs on the current map):
button to preview sample values for the chosen property (this button may not be available if there are too many different values for the chosen property). The table of sample values is sorted by frequency of occurrence (i.e. the number of times each value occurs on the current map):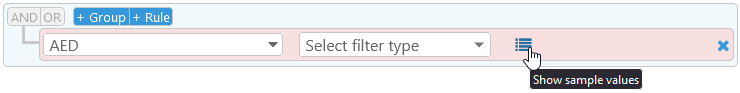
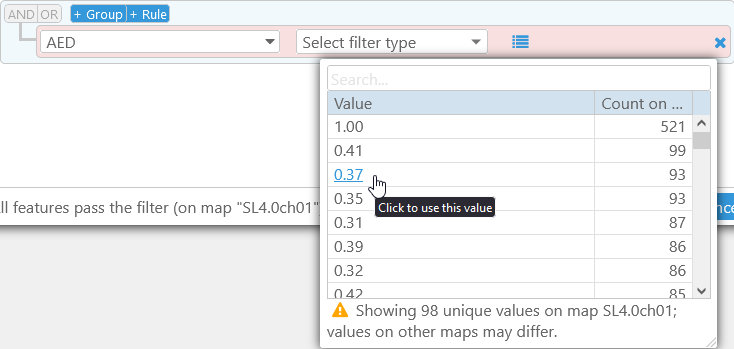
As the warning at the bottom indicates, this table contains only the known values on the current map; values on other maps in the Map set may differ. You can click a value to quickly create a filter that tests for equality:
But you can also manually change the Filter type to some other condition. For example, assuming that the AED qualifier contains a quality score, you can create a filter that will pass through only high-quality annotations (i.e. those with AED > 0.7):
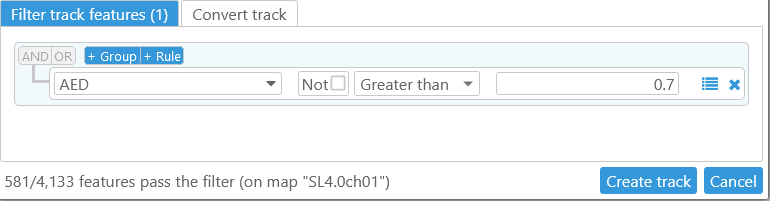
The statistics in the bottom-left corner of the dialog display the number of features that currently pass the filter (depending on the map, this count might take a few seconds to update). When you are satisfied with the filter, click the Create track button to create the virtual track:
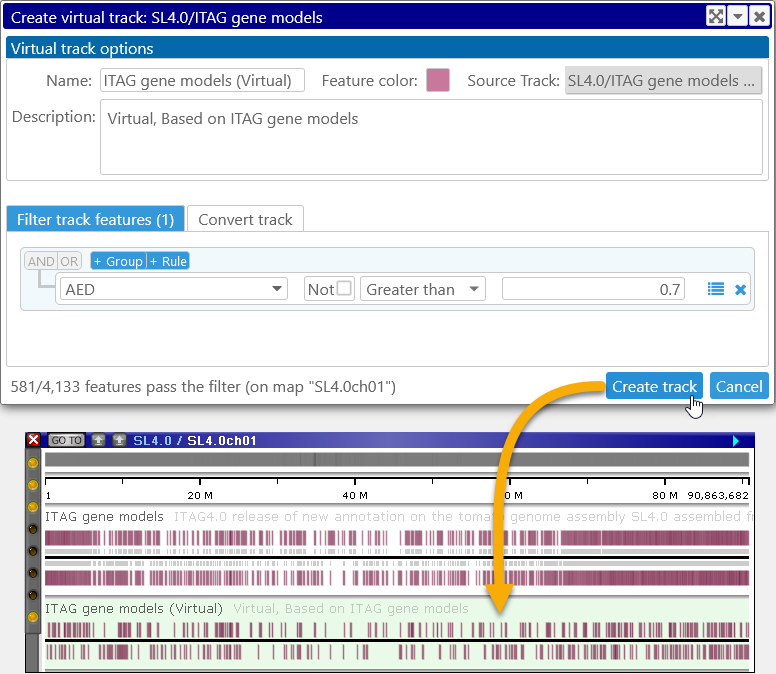
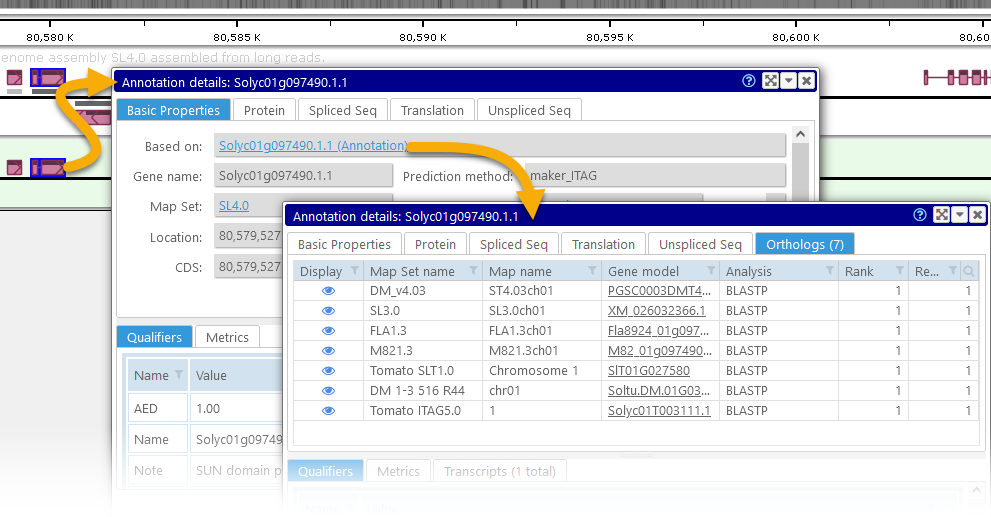
The new virtual track has a light-green background, and contains only the features that pass the filter. As usual, you can zoom in to view individual features, and click a virtual feature (in this case, a virtual gene annotation) to view its properties. Not all of the properties of the original feature are available in this view; for example, Orthologs and Transcripts are not available for virtual annotations. However, the Basic Properties tab page will contain a link to the original track feature; click it to view its full properties:
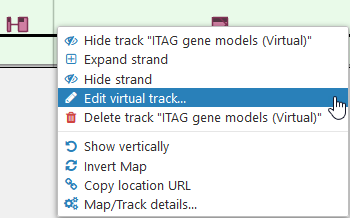
You can edit the appearance of the virtual track (such as its color, name, and description) at any time. To do so, right-click the track, and select Edit virtual track from the popup menu:
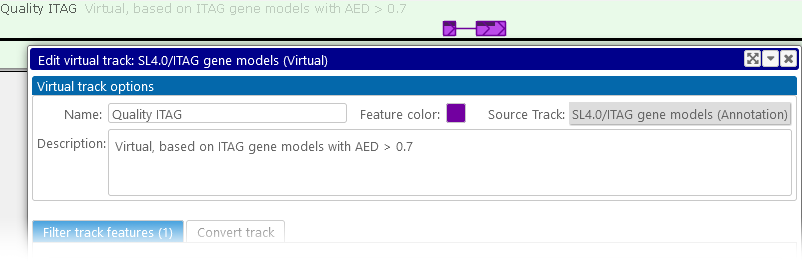
While editing the track, you can also modify existing filtering conditions, add new conditions, and delete conditions (click the  button next to a condition to delete it), then click the Apply changes button to view the results.
button next to a condition to delete it), then click the Apply changes button to view the results.
You can delete the virtual track by selecting the Delete track option from its right-click context menu. All virtual tracks are stored in long-term private storage, but usually take up a negligible amount of storage quota.
Note
Virtual Tracks cannot currently be shared between users; this also means that they will not be shared via bookmarks.
Converting tracks and coloring track features
It is also possible to convert track features from one kind to another (e.g. from gene Annotations to Markers or vice versa). To do so, create or edit a virtual track (as described above), and click on the Convert track tab. Conversion is always applied after filtering, but you can also leave the filtering conditions blank, in which case all features will automatically pass the filter:
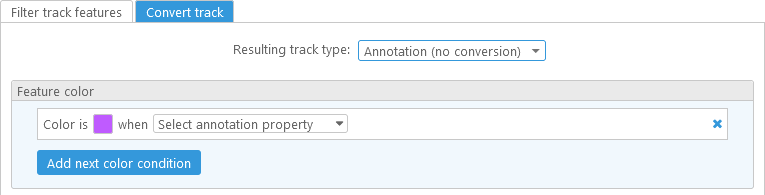
The option that is selected in the Resulting track type dropdown list specifies the transformation that will be applied to the original track features. The no conversion option will preserve the original feature type -- i.e. annotations will remain as annotations, and markers will remain as markers. However, it is possible to re-color track features based on their properties. For example, you can create a track where annotations with high AED are highlighted in green, and those with low AED are highlighted in red:
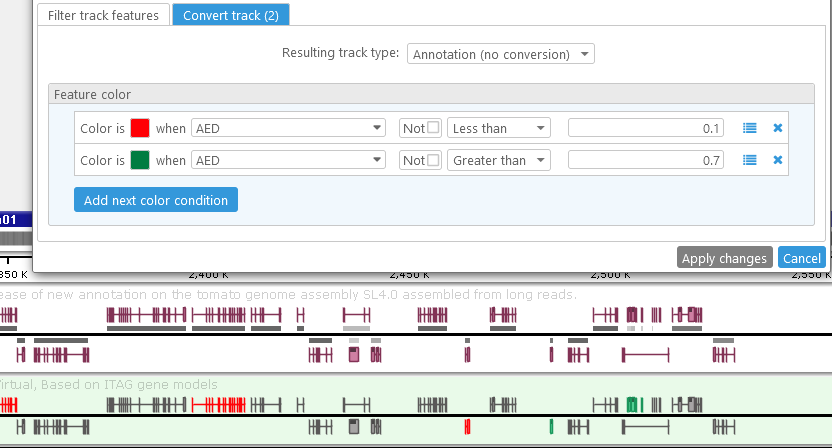
You can also combine track filtering and feature conversion. For example, it's possible to alter the virtual track to hide medium-quality features, and only display the color-coded extremes:
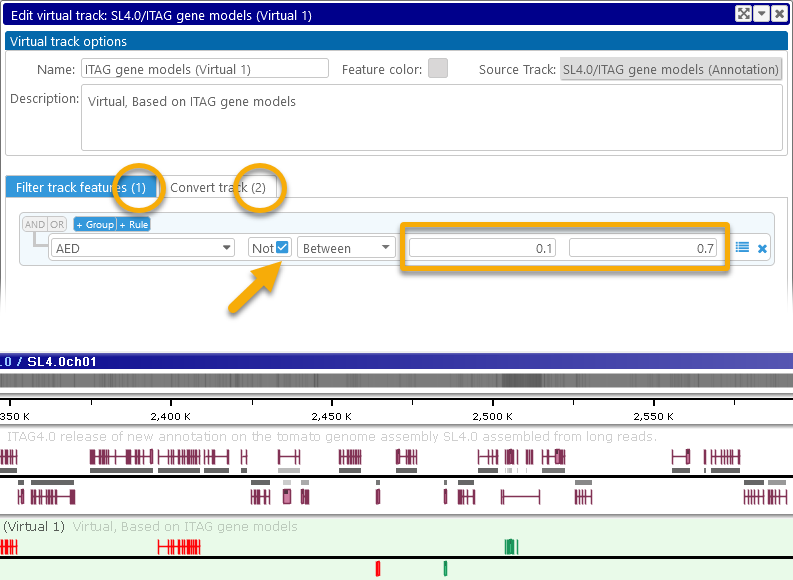
In this example, we checked the Not checkbox to only pass the features who fall outside of the specified numeric range (0.1 .. 0.7). Also note that the number of filtering/conversion conditions is always listed in the tab headers (as shown above); this makes it easy to tell at a glance whether the virtual track applies filters, conversions, or both.
Converting to Markers
The Marker track option in the Resulting track type dropdown list always creates virtual markers based on the original track features. You can specify which properties of the original feature will serve as marker coordinates, marker names, and qualifiers. For example, you could create a marker track based on gene annotations, where the marker labels contain the original annotations' AED scores:
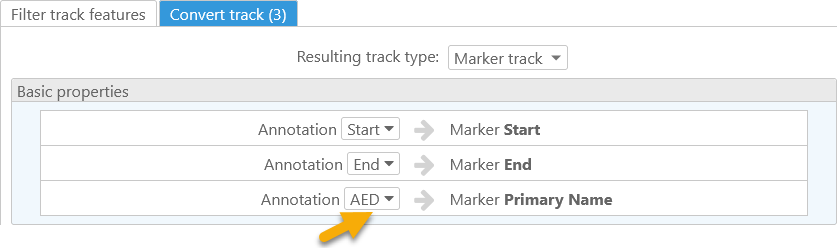
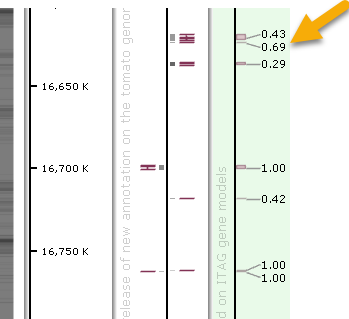
In this example, the map is shown vertically to make marker labels easier to read.
Converting to Annotations
The Annotation Track option in the Resulting track type dropdown list creates virtual annotations based on the original track features. For example, you could create virtual annotations that cover the CDS of the original transcripts:
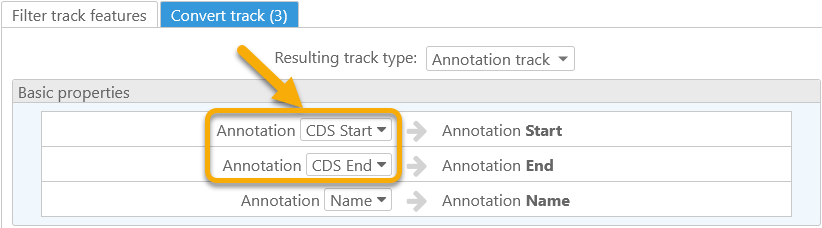
The resulting annotations will always consist of a single exon: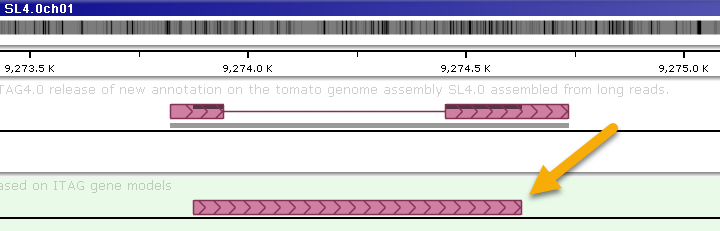
Converting qualifiers
In addition to converting the required basic properties, you can optionally generate qualifiers for the virtual track features: either by copying the original features' qualifiers directly, or by creating new qualifiers based on other properties of the original feature. For example, we can edit the virtual marker track (described above) to add some qualifiers:
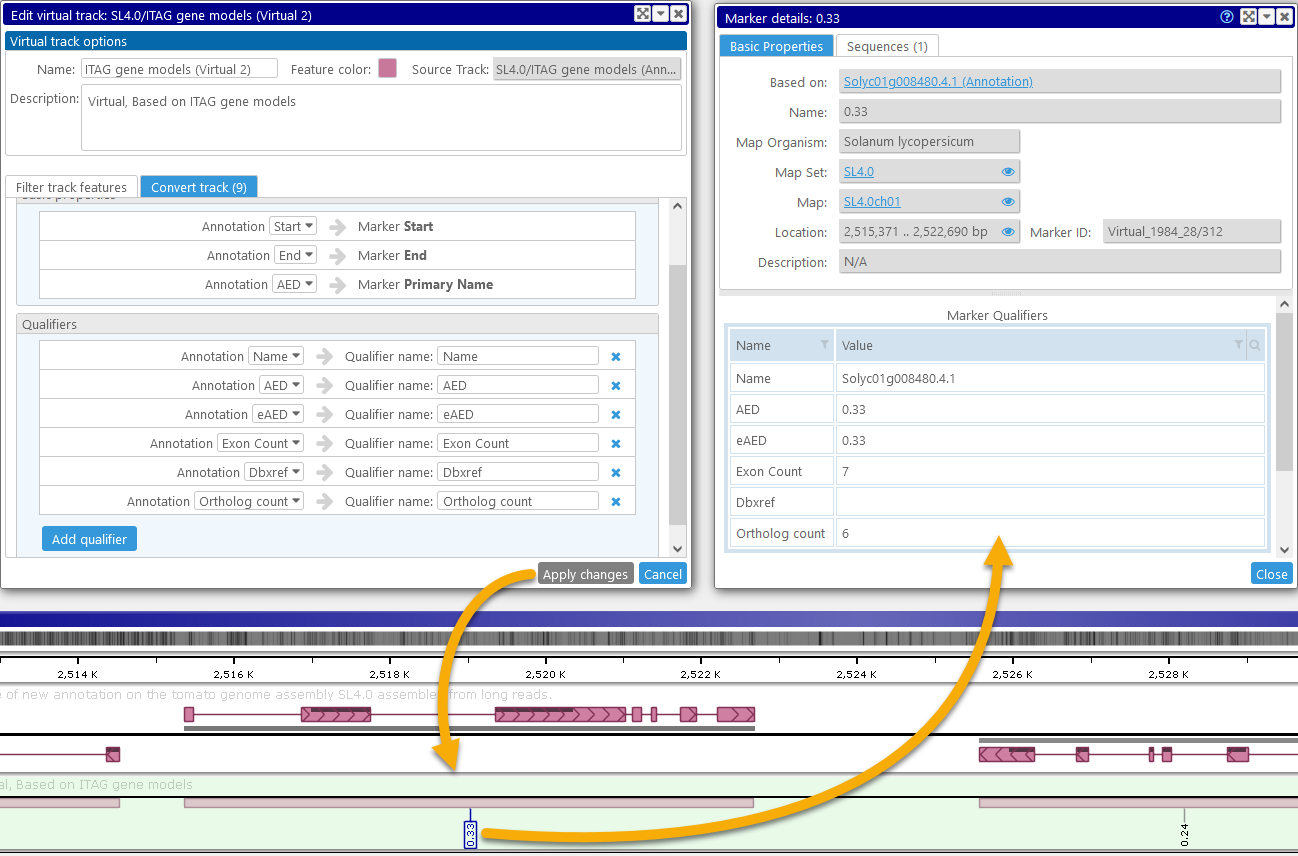
In this example, the "Name" qualifer for the virtual marker contains the original annotation name; the "Exon Count" and "Ortholog count" qualifiers contain the respective calculated properties; and the "AED", "eAED", and "Dbxref" qualifiers are copied from the original gene annotation. The marker label (i.e. its Primary Name) still reflects the AED score (as specified by the original "AED" qualifier).
Filtering with groups
Filtering rules can be combined into groups to create more complex filtering conditions. To create a group, click the +Group button:
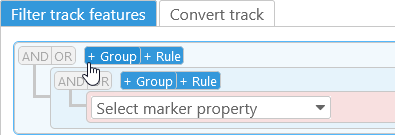
You can then add multiple rules to the group, or create additional nested groups. The AND|OR buttons next to the group determine its behavior: if AND is selected, then a track feature must satisfy all of the conditions within the group in order to pass the filter; if OR is selected, then at least one of the conditions must be satisfied.
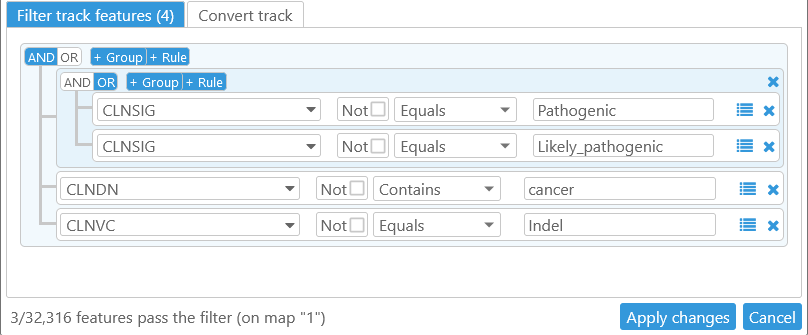
For example, let's create a filter on the ClinVar track (in the Homo Sapiens GRCh38 map set) that identifies indel markers related to cancer:
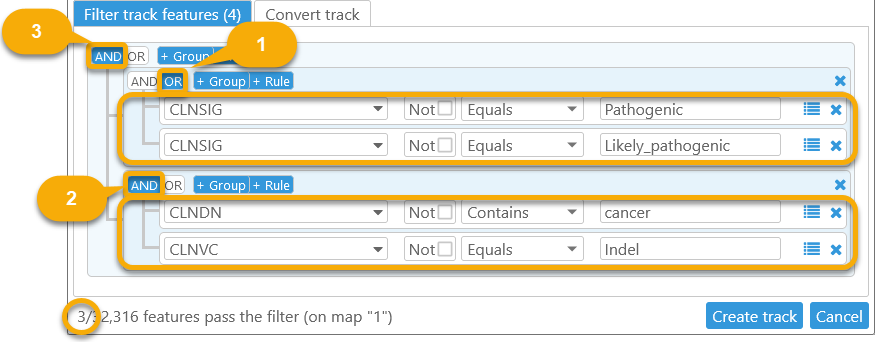
- The first group identifies markers whose CLNSIG qualifier is either "Pathogenic" or "Likely_pathogenic".
- The second group identifies markers whose CLNDN qualifier contains the string "cancer", and whose CLNVC qualifier is "Indel".
- The entire filter combines the two groups, requiring that both conditions must be met.
As the readout in the bottom-left corner of the dialog indicates, only 3 markers pass the filter on chromosome 1: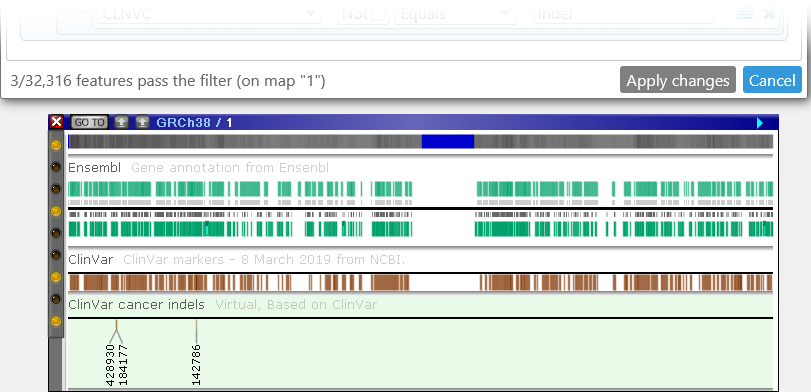
In more formal terms, the condition for this filter can be written as follows:
(CLNSIG == "Pathogenic" OR CLNSIG == "Likely_pathogenic") AND (CLNDN contains "cancer" AND CLNVC == "Indel")
Filter groups can be nested, and can contain a mixture of rules and other groups. For example, the filter above can be simplified as follows:
(CLNSIG == "Pathogenic" OR CLNSIG == "Likely_pathogenic") AND CLNDN contains "cancer" AND CLNVC == "Indel"