Web Persephone: The Marker Details dialog
This dialog displays all known properties of a marker, including its qualifiers, known locations, and sequences (if any). To open the dialog, click the marker on the Marker track (or anywhere else in Persephone, e.g. in the Search results or Export tables). If the marker's track is visible, the marker you clicked will be outlined with a solid border:
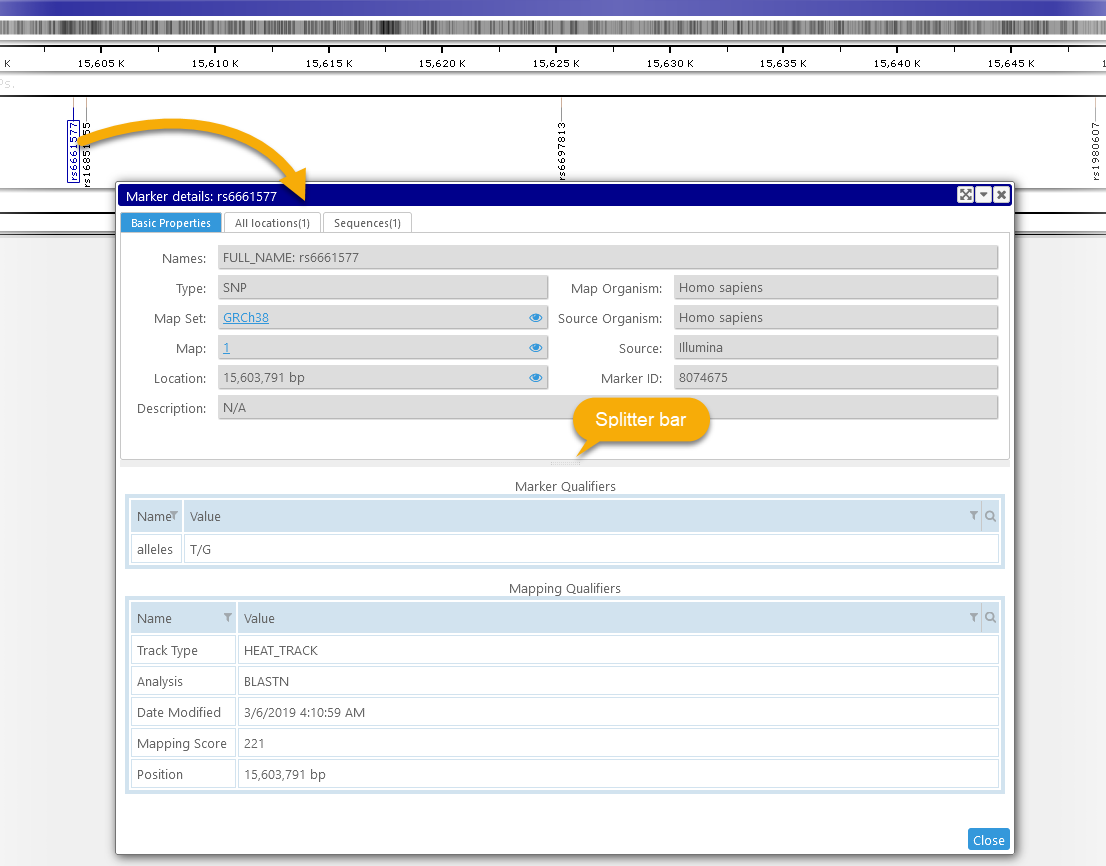
The marker's properties, locations, and sequences are displayed in separate tabs in the top pane of the dialog; and its qualifiers are displayed in the bottom pane. As usual, you can drag the splitter bar to resize the panes. By default, the Basic Properties tab will be selected; however, the dialog will "remember" which tabs were selected the last time it was opened, and select the same tabs the next time it is opened.
Top: Basic Properties
This tab displays all the basic metadata that is known about the marker.
|
Names: |
Lists all the names for this marker. While most markers will only have one name, others can have multiple names, e.g. representing the marker's identifiers in various external databases. |
Map organism: |
The organism of the current map. |
|
Type: |
The marker's type (multiple markers on the same track may have different types). |
Source organism: |
The organism where the marker originally came from; this may be different from Map organism. For example, a marker in Zea mays may later be mapped to Sorghum bicolor. |
|
Map set: |
The map set where this marker is located. Click the map set name to open its Map set details dialog; click the |
Source: |
The origin of this marker; typically, the name of the organization who produced it. |
|
Map: |
The map where the marker is located. Click the map name to open the Map details dialog; click the |
|
|
|
Location: |
The start coordinate (and the end coordinate, if any) of the marker on the map. Click the |
|
|
|
Description: |
Detailed description of the marker (if any). |
|
|
Top: All locations
A marker may be mapped to multiple locations, either within the same map set (and even on the same map), or to locations in different map sets (e.g. across different organisms). The All locations tab displays all known locations of the marker:
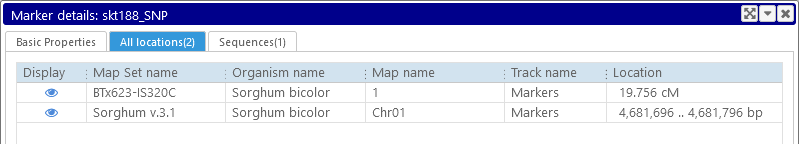
Note that the location of the marker may be displayed in either bp (on physical maps) or cM (on genetic maps). Click the  icon to open one of the maps where the marker occurs; highlight the marker; and zoom into its location:
icon to open one of the maps where the marker occurs; highlight the marker; and zoom into its location:

The marker will be automatically connected to its location on the current map by a line connector.
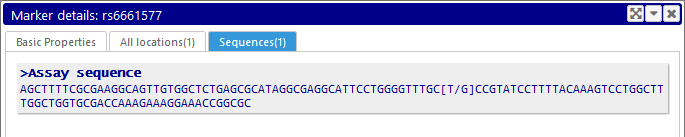
Top: Sequences
If the marker has any sequences associated with it -- such as the assay sequence, primers, etc. -- they will be displayed on this tab in FASTA format. Otherwise, the tab will be hidden.
Bottom: Qualifiers
Each marker can have two set of qualifiers (i.e., key-value pairs) associated with it. The Marker Qualifiers describe the marker itself; whereas the Mapping Qualifiers describe the reason why the marker was mapped to this particular location (as well as other metadata about this mapping).
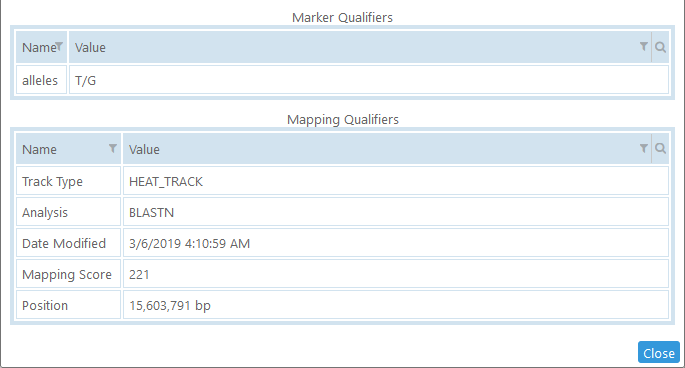
Just like annotation qualifiers, marker qualifiers may be formatted as links (leading to external databases or Web-based tools).
Note
The table of qualifiers is equipped with the standard table sorting and filtering controls.
