Web Persephone: Annotation Metrics view
This view (accessible from the Annotation Details dialog) displays detailed measurements of the annotation's sequence. By default, the view shows the annotation and its exons:

Drag the mouse across the view to select a portion of the sequence and display all of its measurements:
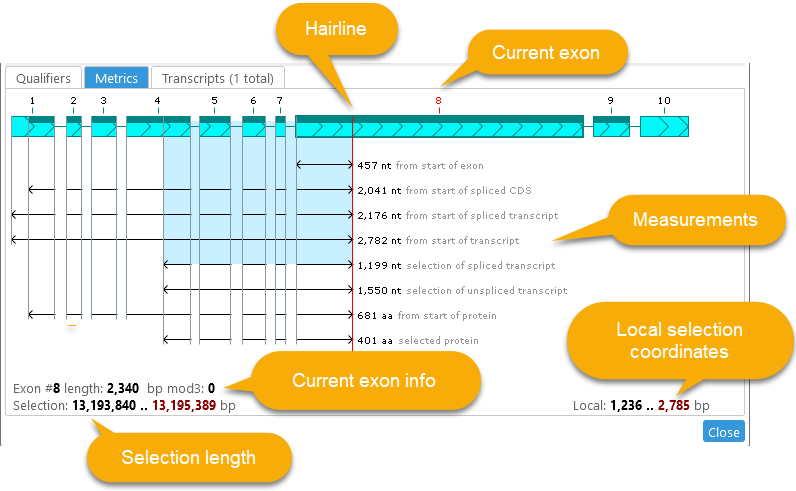
The red "hairline" shows the current location of the cursor; the current exon number is likewise highlighted in red. Detailed measurements are shown underneath the annotation's model, e.g. the total length of the currently selected sequence, the offset of the selected sequence from the start of the current exon, etc. Local selection coordinates (i.e., relative to the start of the annotation) are displayed in the bottom-right; absolute selection coordinates are displayed in the bottom-left, in addition to the length of the current exon and its offset modulo 3.
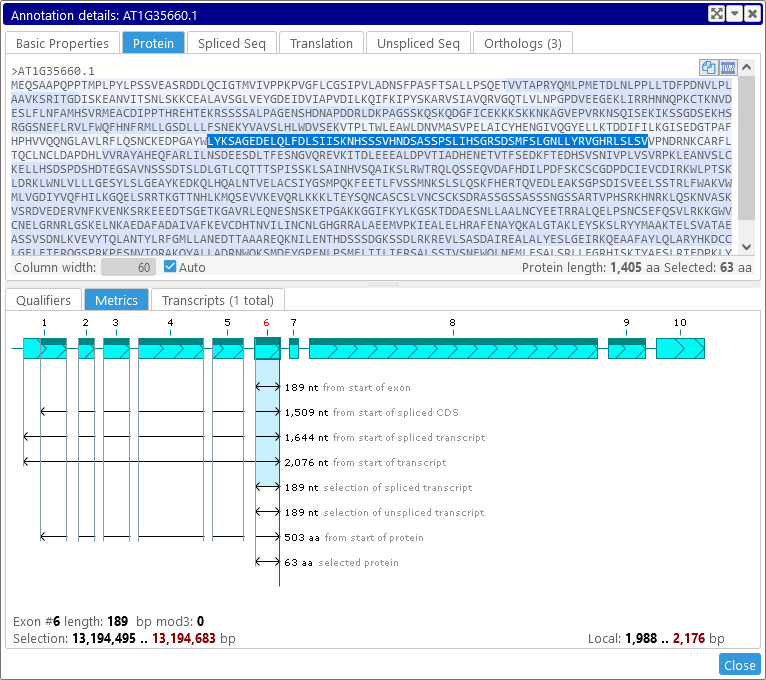
Note that graphical selection in the Metrics view is synchronized with textual selection on the sequence tabs of the Annotation Details dialog. For example, if you select the Protein tab, you can see which aminoacids are represented by the currently selected sequence:
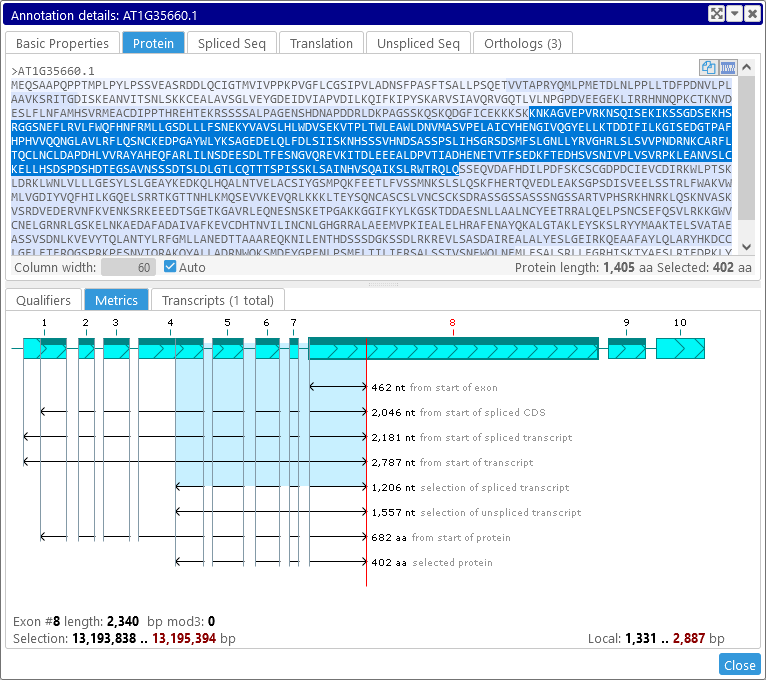
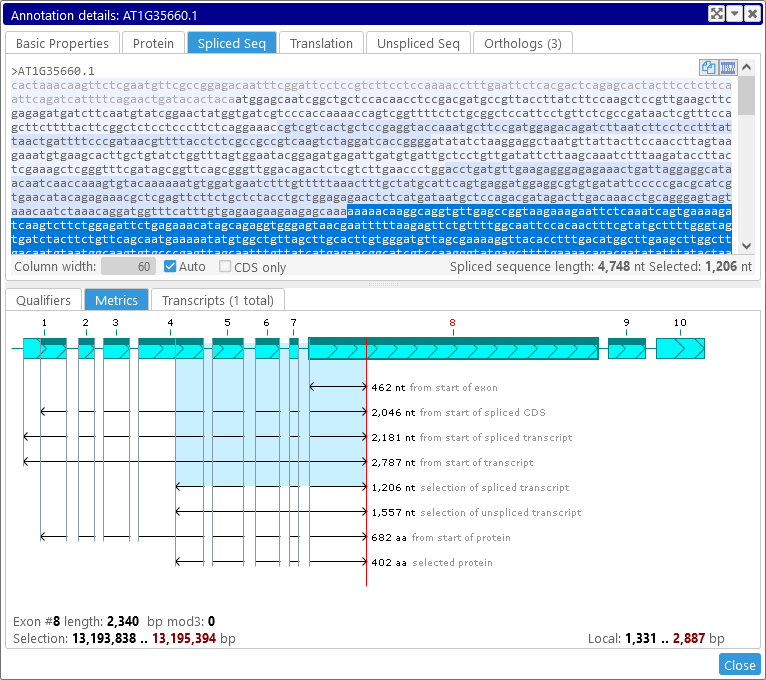
Selecting the text in the Protein tab will affect the graphical selection in the Metrics view, and vice-versa. The same applies to the Spliced Seq and Unspliced Seq tabs; for example:
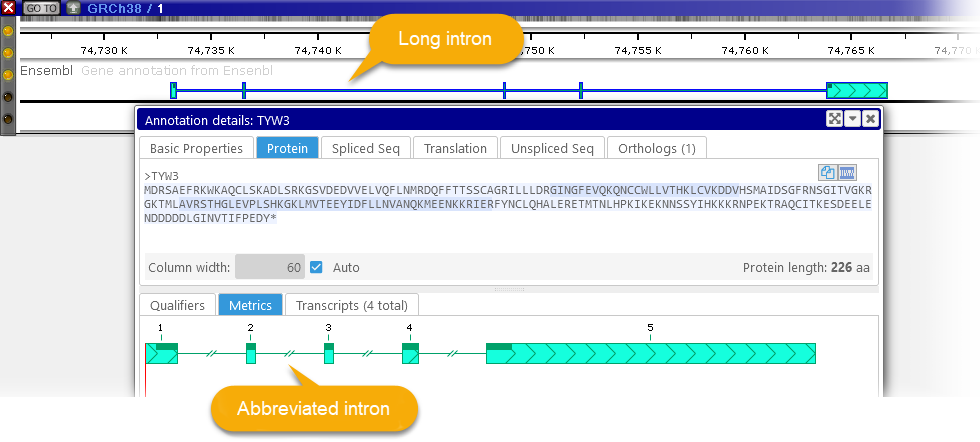
Note that especially long introns (e.g. in transcripts on Homo Sapiens) may be abbreviated in the Metrics view:
Precise navigation
You can double-click an exon to quickly select it:
You can also use the Left arrow / Right arrow keys on your keyboard to move the cursor by exactly one nucleotide. Hold down the Shift key while pressing the arrows to select the sequence as the cursor is moving (in a way similar to selecting text). Hold down the Ctrl key and press the arrows to "jump" the cursor to the nearest boundary: the start/end the nearest exon, or the start/end of the CDS.
