Web Persephone: Import sequences
Sample file: https://persephonesoft.com/data/import/at.1-2.fna.gz
You can import a file in the FASTA format to create a new map set. The file may contain multiple records, with record headers starting with '>' and ending with a newline; for example:
>NC_003070.9 chromosome 1 sequence
ccctaaaccctaaaccctaaaccctaaacctctGAATCCTTAATCCCTAAATCCCTAAATCTTTAAATCCTACATCCATG
AATCCCTAAATACCTAAttccctaaacccgaaaccggTTTCTCTGGTTGAAAATCATTGTGtatataatgataattttat
CGTTTTTATGTAATTGCTTATTGTTGTGtgtagattttttaaaaatatcatttgagGTCAATACAAATCCTATTTCTTGT
GGTTTTCTTTCCTTCACTTAGCTATGGATGGTTTATCTTCATTTGTTATATTGGATACAAGCTTTGCTACGATCTACATT
TGGGAATGTGAGTCTCTTATTGTAACCTTAGGGTTGGTTTATCTCAAGAATCTTATTAATTGTTTGGACTGTTTATGTTT
...
>NC_003071.7 chromosome 2 sequence
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
...
After loading the file, Persephone will display all the record headers found within it, and will attempt to automatically extract map names and chromosome names:
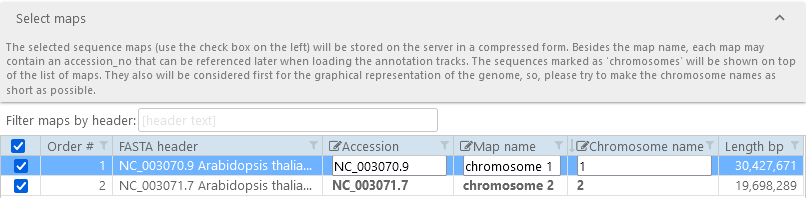
By default, all records are checked; however, you can un-check any records that you do not wish to import (e.g. to save space, or to reduce clutter).
Click on a map record to manually edit its accession, map name, and chromosome name. Chromosome names are especially important if the imported file contains a mix of chromosomes and scaffolds; maps with non-empty chromosome names will always be shown on top of the map list, and will be given priority when the entire genome is shown graphically. To manually edit one of the map's properties, type the new value into the corresponding textbox; for example:
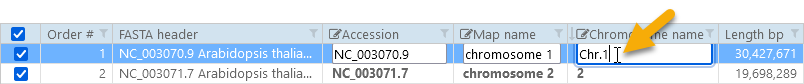
Alternatively, you can select multiple records, then click one of the  buttons to edit their corresponding properties in bulk (as usual, Ctrl-Click a record to toggle its selection individually; Shift-Click to toggle selection for a block of records, or press Ctrl-A to select all records):
buttons to edit their corresponding properties in bulk (as usual, Ctrl-Click a record to toggle its selection individually; Shift-Click to toggle selection for a block of records, or press Ctrl-A to select all records):
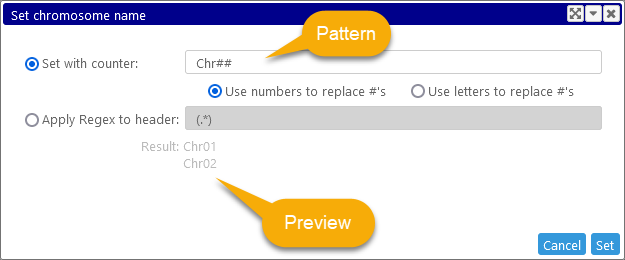
By default, you will be prompted to label the records sequentially. Any '#' characters in the pattern will be replaced by either sequential numbers (1, 2, etc.), or letters (AA, AB, etc.), depending on your choice. The gray result preview text will change to display a sample of the resulting values.
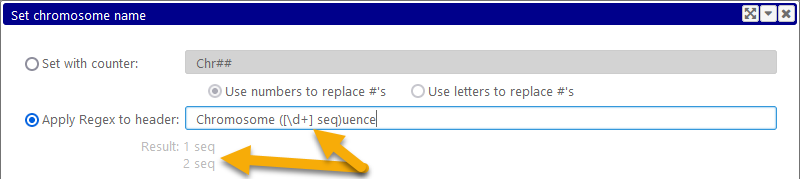
Alternatively, you can click the Apply Regex to header option. In this case, the pattern will be interpreted as a Regular Expression, and applied to the entire record header in order to extract its relevant portion. Only the portion of the text that is matched by the first group (the sub-pattern enclosed in parentheses) will be extracted. For example:
When you are done editing map names, click Next to set the properties of the imported map set: its organism, name, and description.
Click Next again to index the imported maps for BLAST (this process may take some time). The new map set will be displayed in the map set tree in purple, indicating that it is stored in long-term private storage rather than the main Persephone database:
You can now view the imported sequences; run BLAST queries against them; or import additional tracks and attach them to these maps.
