Web Persephone: The Local Variants dialog
To open the Local variants dialog, click a SNP (or an indel) on the Variants track:
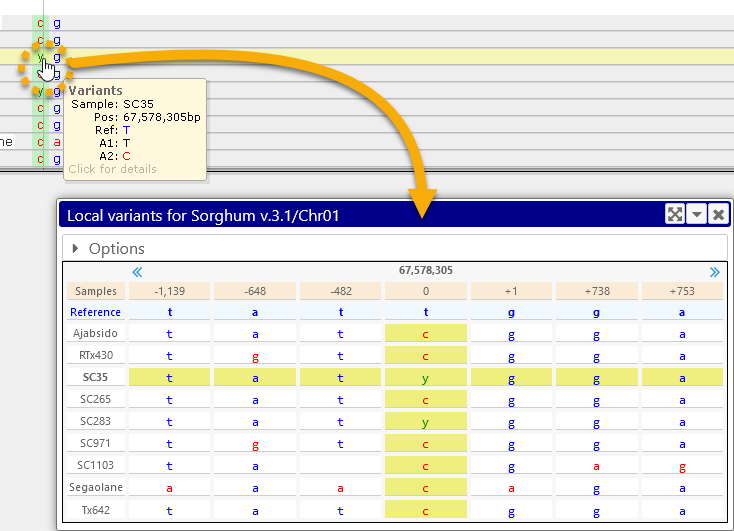
This dialog displays all SNPs in the current "neighborhood" as a table. By default, samples are listed horizontally as rows and variants are listed as columns (just as they are on the track). The current variant is highlighted with a yellow background; its location on the track is also highlighted with a light green background. You can click a table cell to select a different variant (you can also click the name of a sample to highlight that sample). The newly selected variant will scroll into the central position in the table, and the map will likewise scroll to this variant (assuming it is off-screen):
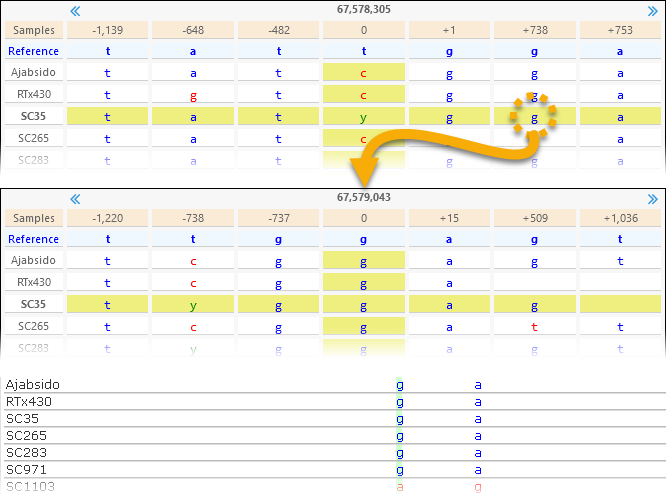
Likewise, you can click a different variant on the map to display it in this dialog.
The Navigation row at the top of the dialog shows the absolute position of the current variant and relative positions of its neighbors. Click the pagination buttons at the top of the bar to quickly move to the previous/next variant:
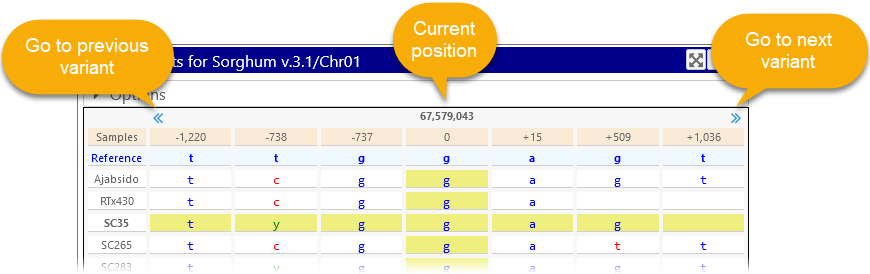
The Reference row rests directly below the Navigation row, and displays the reference sequence allele at the current position.
Note
The Reference row always shows the reference sequence, even if the coloring schema is set to Reference genotype or Two parents.
Display options
Click the Options button to expand the Options panel:
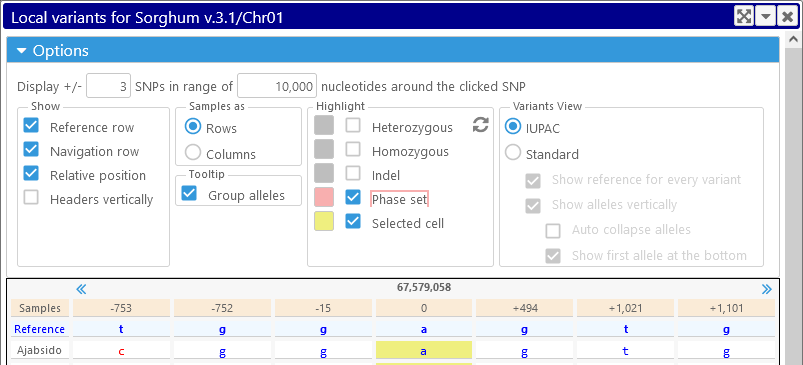
The top row of the Options panel controls the number of variants (i.e. SNPs or indels) that should be shown around the currently selected position. For example, you could opt to display 5 variants to either side, but only if they're no further than 800 bp away:
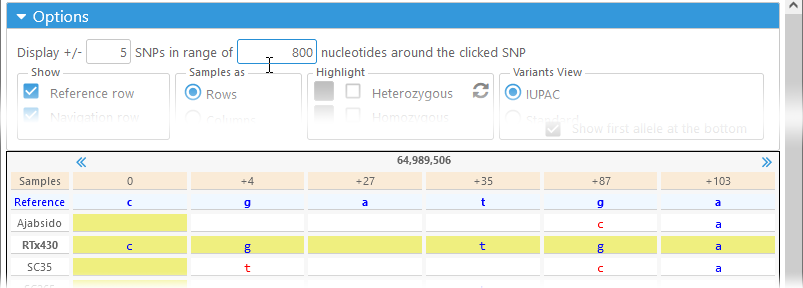

In this case, the currently selected variant is shown on the left side of the table, since the variants upstream of it are further than 800 bp away.
The rest of the options in this panel are organized into several logical groups, as described below.
Options group: Show
- Reference row: shows or hides the Reference row in the table header.
- Navigation row: shows or hides the Navigation row in the table header.
- Relative position: displays variant positions as offsets relative to the position of the currently selected variant. This is the default; uncheck this checkbox to display absolute coordinates for every variant.
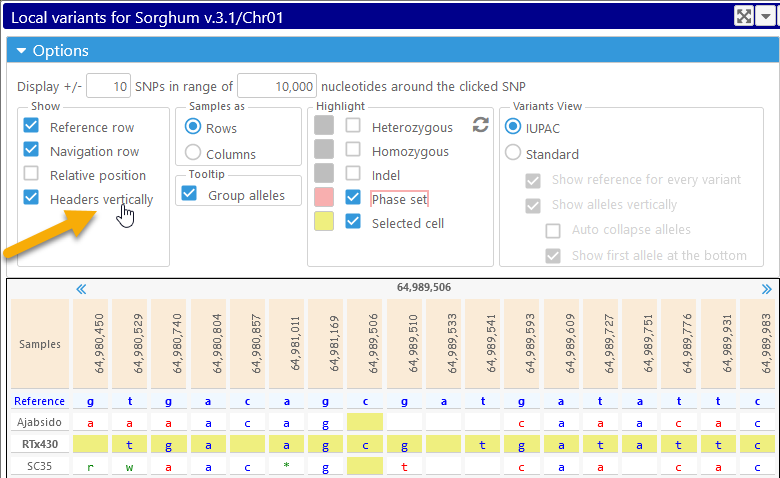
- Headers vertically: displays variant positions vertically. This can be useful when displaying absolute coordinates, to prevent them from overlapping:
Options group: Samples as / Tooltip
- Samples as Rows/Columns: rotates the entire table 90 degrees, to display the variants vertically:
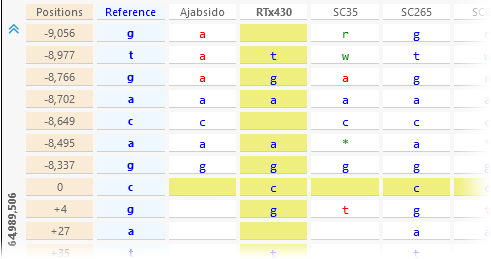
This can be useful if the map is displayed vertically in the main view. - Group alleles: changes the way alleles are shown in the mouseover tooltip. If this option is checked, heterozygous indels are grouped by their reference; otherwise, they are all shown inline:
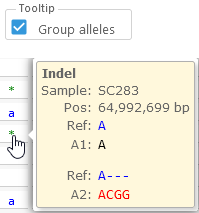
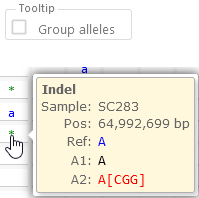
You can use this option to reduce clutter when examining polyploid variants.
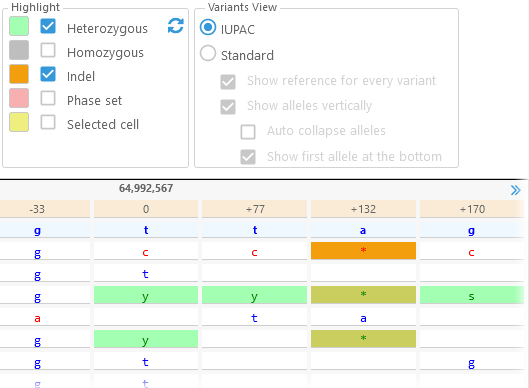
Options group: Highlight
These options control highlighting of specific kinds of variants. Click the color swatch next to each option to set your preferred highlight color. For example, you could choose to highlight indels in orange, and heterozygous variants in green; note that these highlight colors will overlap for indels that are also heterozygous:
Options group: Variants view
These options control the way variants are printed in the table.
- IUPAC: By default, variants are printed using IUPAC codes. For example, a diploid heterozygous SNP whose alleles are C and T will be printed as "y".
- Standard: displays every allele in detail (similarly to how it's listed in the VCF file).
- Show reference for every variant: If this option is checked, then every variant is prefixed by the reference allele. This is especially useful when viewing indels. For example, you could tell at a glance which nucleotides were deleted for this variant:
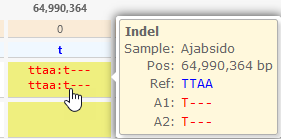
- Show alleles vertically: Uncheck this option to print alleles on the same line, separated by a slash, similarly to how they are listed in the VCF file:

- Auto collapse alleles: Check this option to show only the distinct alleles for heterozygous SNPs/indels; by default, every allele is shown.
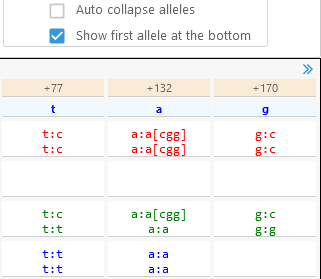
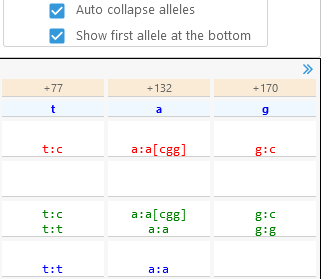
This option is useful for reducing clutter when viewing polyploid variants. - Show first allele at the bottom: Reverses the order in which alleles are displayed in the table.
