Control Files
The PersephoneShell command add uses the INI-format control files to perform many important tasks, including setting parameters and loading data from local and remote files. INI files are simple text files with a basic structure composed of sections, properties, and values.
Note
For more details about the INI file format, please see the Wikipedia article "INI file". Note that the INI file format is loosely defined, so we extended it a bit. For example, we allow multi-line values enclosed in double quotes.
You can use control files from the installed directories (see Sample Control Files). In addition, you can modify and create control files with a plain text editor (see Creating and Modifying Control Files). Please note, if you plan to modify or create control files, the data source formats vary depending on the object type (e.g., annotation, organism). See Data Source Formats Supported for more information.
Sample Control Files
The PersephoneShell package includes a folder called Samples that has many sample control files. For example, the Samples/Organism folder includes the organism control file "Beta_vulgaris.ini", which can be used by the add command to add the organism Beta vulgaris subsp. vulgaris (sugar beet) to your Persephone database. It is very common to create new INI files by copying the existing ones and modifying few values. Find a proper folder for the given data type: Organism, Sequence, Annotation, etc. Sometimes, the sample file name contains the name of the organization producing the data. For example, the format of the FASTA headers or a set of GFF attributes is common for the data source, such as NCBI or Phytozome. The parsing logic for different data sources is preset in the sample files that have the source organization at the end of the file name: beetEL10-ncbi.ini, IRGSP-1.0-gramene.ini. Find a template best matching your data source and create a copy of the INI file. This will minimize your typing and will save you quite some time.
Creating and Modifying Control Files
You can use a standard text editor to edit the control files. It is highly recommended using the editors that support the INI file syntax highlighting (Notepad++, vim, mcedit).
If you wish to modify existing or create new control files, please keep the following in mind.
- Refer to the Wikipedia article "INI file" before you begin.
- Use existing control files as reference, templates, or both.
- Read the detailed comments (lines that begin with a semicolon [;]) in existing control files for guidelines and syntax rules.
- Most of the Section (e.g., "[MapSetTree]") and all property (e.g., "FileType") names are pre-defined and cannot be modified.
- An ID value (e.g., "MapSetId") is a 64-bit unsigned integer.
- A Boolean key (e.g., "IsSearchable") can only be set to true or false. Other values such as Y/N or 0/1 are not allowed.
- A string value with blank spaces that contains line breaks should be enclosed in double quotes (" "). To enter a double quote mark in a string put a slash (\) before it (e.g., "the common name is \"Sugar Beet\"."). One-line values can be specified without double quotes.
- When multiple values are assigned to the same key, list them on separate lines. If one of the values (e.g., a URL) contains the equal sign ('='), the entire multi-line text must be enclosed in double quotes:
Dbxref="https://www.ebi.ac.uk/interpro/entry/InterPro/%s/|InterPro:(IPR\d+)
https://www.ebi.ac.uk/interpro/entry/pfam/%s|Pfam:(PF\d+)
http://www.pantherdb.org/panther/familyList.do?searchType=basic&fieldName=all&organism=all&listType=6&fieldValue=%s|(PTHR[^,\s]+)" - The control file must be saved with an ".ini" extension. Other extensions (e.g., ".txt", ".bat") are not supported.
- The data files with actual data are referenced in the INI file using Source or Sources keyword. Source can point to a single file or to a directory, in which case all files from the directory will be processed. The files can be compressed with gzip and have extension ".gz". Particular formats are listed for each of the object types.
- The Description fields allow some simple HTML tags like <b> for bold, <i> for italic, <a> for hyperlinks. Please remember to "escape" the quotation marks around the URL in the hyperlink <a> tag using back slash:
Description="The data downloaded from <a href=\"ftp://ftp.ncbi.nlm.nih.gov\">NCBI</a>".
The map's or map set's description is shown in the corresponding Properties window. The track description can also be shown in a tool tip that appears over the track panel on mouse over. The long multi-line text for this tool tip is trimmed, only the first line is shown: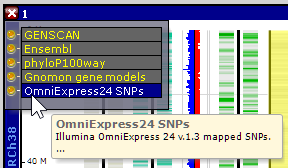
Data Source Formats Supported
The table below lists data source formats supported by PersephoneShell. Click the object name for sample control file sections that show how the data source is loaded. Please note, comments have been removed for brevity.
|
Objects |
Data Source Formats Supported |
|
FASTA, GenBank |
|
|
GFF, GTF, GenBank, BED |
|
|
Sequence Database |
GenBank |
|
VCF |
|
|
GFF, GVF, TXT (CSV, TSV) |
|
|
Command lines in the control file |
|
|
TBLASTN |
|
|
GFF, CHAIN, TXT, PAF |
|
|
bedGraph, bigWig |
|
|
TXT |
|
|
TXT |
|
|
TXT |
Common sections
[ProcessRun]
; RunId: if specified, it uses the existing run.
;RunId=12345
; Run description: if specified, a custom description will be used. Will be ignored
; if a RunId is specified.; otherwise, "Added annotations for {MapSet Accession No.} from {Sources}." will be used.
RunDescription="Loaded annotations for BrapaFPsc_v1.3 from http://phytozome.jgi.doe.gov/pz/portal.html#!bulk?org=Org_BrapaFPsc"
A Process RUN can be considered as a batch job with an ID and description. The objects inserted during one run can later be deleted together using 'delete run' command. Each loading process creates a record in the PROCESS_RUN database table. If you plan to load several sets of data that should be considered as part of one job, for the first load you need to provide description of the job and reference the newly-created RunId in the later sessions. The subsequent loads of logically-linked related data should specify the existing RunId using RunId= construct. In this case the RunDescription field should be commented out, and RunId, that is common for the parts of the data, should be used. PersephoneShell can list existing RUNs with 'list runs -l' command. The switch '-l' is used to display extra fields in a 'long' output format with a column header.
[MapSet]
; Either MapSetId or MapSetPath is required.
; MapSetId: id of a target map set.
;MapSetId=247848026
; MapSetPath: path of a target map set.
MapSetPath="/Brassica rapa/BrapaFPsc_v1.3"
Tracks with the data of different types are added to the maps of an existing map set. The map set can be referenced by MapSetId (a numeric identifier) or by MapSetPath (a literal path that includes location of the map set in the map set tree).
Note
You can use PersephoneShell to help yourself with typing the full path to the map set. For example, type 'list maps' on the prompt and then (after adding a space) start typing the first root node of the branch that leads to the map set. For example, if full path of map set called TAIR10, is "/Arabidopsis thaliana/TAIR10", you can start typing 'list maps /Ara', then press TAB to auto-complete "/Arabidopsis thaliana", then type '/'. Next, pressing TAB would cycle through all map sets in '/Arabidopsis thaliana' node, one of them should be 'TAIR10'. Copy the resultant map set path together with the double quotes and paste it into the control file. You can also use the command 'list path'.
To get MapSetId, you can use 'list mapset -l' command. Again, using '-l' would force PersephoneShell to use the long output format that prints the column header of the table.
Note
You can use '-p' command option with a filter pattern to limit the list of map sets. For example, to show info about 'TAIR10' you can type 'list mapset -p TA*'
[MapMapping]
; If no mapping is found in this section, it is assumed that each MAP_NAME in file exactly matches a MAP_NAME in DB.
; If map names in file are different from those in DB, map each MAP_NAME in file to a MAP_NAME in DB.
; Otherwise, annotation won't be added.
; MapsIdentifiedBy: one of MapId, AccessionNo, GenomeDnaId, MapName (default)
;MapsIdentifiedBy=GenomeDnaId
; LoadListedMapsOnly: if true, only data for the maps listed in this section will be added.
; If false, PersephoneShell will still try to match names from the file to maps in the database
; using MAP_NAME and will report an error if no match is found
LoadListedMapsOnly=true
;MAP_NAME in file=MAP_NAME in DB
83=chromosome 1
84=chromosome 2
85=chromosome 3
86=chromosome 4
87=chromosome 5
88=chromosome 6
89=chromosome 7
90=chromosome 8
91=chromosome 9
92=chromosome 10
It is quite common when an annotation file uses names for the sequences that are different from the names in the database. In this case, you should create a name translation table in the MapMapping section. For each sequence name in the file provide MAP_NAME of corresponding map in the database. If the file references a map that is not listed in the table, depending on the value of the flag LoadListedMapsOnly, the data lines will be skipped (true) or a fall-back procedure of finding maps without the translation table will be used, and if a matching map is not found, an error will be reported (see the comments in the code above).
Alternatively, if you know that the maps are identified by their MapId, AccessionNo or GenomeDnaId, you can use an instruction telling that the names in the file are some specific kind of identifier:
MapsIdentifiedBy=GenomeDnaId
The marker mapping files can be produced by running BLASTN. The file format for the BLAST library files contain GenomeDnaId - a number identifying genomic sequences in the database. The tabular output format of BLASTN would contain the subject sequence identifiers (GenomeDnaId) and it would be convenient to use the instruction above telling PersephoneShell that the column with map names actually are GenomeDnaIds. It this case, the name translation table would not be needed.
Sources
The majority of the control files require corresponding data files. The sources can be represented by a single file:
Sources="$DATA/Genome/Plants/Oryza sativa MSU_osa1r7/all.con.fa"
or by all files in a given directory:
Sources="$DATA/Genome/Plants/Oryza sativa MSU_osa1r7/sequences"
You can list the files one after another:
Sources="$DATA/arabidopsis/chrom1.fa","$DATA/arabidopsis/chrom2.fa","$DATA/arabidopsis/chrom3.fa","$DATA/arabidopsis/chrom4.fa","$DATA/arabidopsis/chrom5.fa"
If a file has extension .gz or .bz2, it will be automatically uncompressed on the fly. Compressed tar files will be also properly handled if the file names have extensions .tar.gz, .tgz or .tar.bz2.
Remote files can be specified by URL using protocols http, https or ftp:
Sources="ftp://ftp.gramene.org/pub/gramene/CURRENT_RELEASE/data/fasta/oryza_sativa/dna/Oryza_sativa.IRGSP-1.0.29.dna.nonchromosomal.fa.gz"
Note
Some http or ftp servers do not support the stream seek operation needed for some loading procedures. In this case, if you notice problems reading the remote files, please try to download them and use the local copy.
