Web Persephone: Linking into Persephone
Persephone provides a simple Web API for linking to specific locations or features and opening external files. For example, you could construct a URL to link to a specific marker from an external application, or to import an external data file. In general, all such URLs take the form of a standard URL query string; for example, "https://web.persephonesoft.com/?searchAnnot=AT1G20180.1".
Specifying a map set
Most Persephone URLs options allow you to specify a map set to link to (and some options require a map set). To do so, add a URL parameter of the form:
data=genomes/{map set accession}
For example, "data=genomes/TAIR10" links to the TAIR10 map set of Arabidopsis. You can find the map set accession in the Map set details dialog:

Link to a location on a map
This URL option causes Persephone to open a map, and zoom into a specific region. You can copy such a link directly from the map's popup context menu, or you can construct it manually like so:
?data=genomes/{map set accession}&loc={map name}:{start coordinate}..{end coordinate}
Specifying the map set is required for this option. For example, this link will open Chromosome 1 of Arabidopsis, and display the region between 1M bp to 2M bp:
https://web.persephonesoft.com/?data=genomes/TAIR10&loc=1:1000000..2000000
Search for an annotation by name
This URL option will search for an annotation by name:
?searchAnnot={annotation name}
You can optionally specify the target map set, as described above. Upon opening this URL, Persephone will search for an annotation by name (optionally restricting the search to the specific map set). If exactly one annotation (or gene locus) matches the query, Persephone will automatically display the search result (by opening its map, displaying its track, zooming to the annotation's region and highlighting it). For example, the following URL links to the AT1G07390.2 annotation in Arabidopsis:
https://web.persephonesoft.com/?searchAnnot=AT1G07390.2
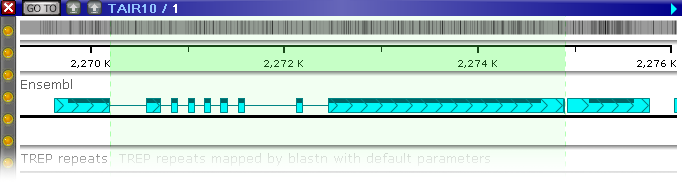
In this case, the annotation is one of the alternative isoforms for the gene; as usual, you can expand the track to show all the isoforms (including the AT1G07390.2 annotation):

You can also use wildcards in your query. For example, you could search for all annotations whose name starts with "AT1G07":
https://web.persephonesoft.com/?searchAnnot=AT1G07*
In this case, Persephone will display the standard search results dialog listing all of the matching annotations:

Search for a marker by name
This URL option is similar to searching for an annotation, as described above:
?searchMarker={marker name}
As usual, you can use wildcards in your query; you can also optionally specify a map set.
Import an external file
This URL option imports an external file into Persephone. Currently, only BAM/CRAM and BED files can be imported directly from the URL, although support for other file formats is currently under development.
To import a BAM/CRAM file, specify the URLs to both the .bam and .bai files (or the .cram/.crai files) separated by a semicolon (";"). Additionally, you must specify the target map set (as described above):
?files={http://url/to/file.bam};{http://url/to/file.bam.bai}&data=genomes/{map set accession}
To import a BED file, specify its URL as well as the target map set:
?files={http://url/to/file.bed}&data=genomes/{map set accession}
As usual, you can provide raw as well as compressed files (e.g. "http://url/to/file.bed.gz").
The import process will begin in the background; when it is finished, Persephone will open the first map from the file and display it. For example:
https://web.persephonesoft.com/?files=https://web.persephonesoft.com/bio/bed/GLEAN.bed&data=genomes/PGSC_DM_v4.03

Files that are imported using this method always be loaded into the local browser session (as indicated by the pink background color of the track); they will disappear when you close Persephone's current browser tab. Naturally, you can always manually import these files into long-term private storage by using the standard Import dialog.
Note
When linking to external files, make sure that the map names in the file exactly match the map names in the target map set. For example, if maps in the external file are named "chr.1", "chr.2", etc; and maps in the target map set are named "1", "2", etc.; then the import process will fail.
Open a bookmark
This URL option opens a bookmark:
?bookmark={bookmark ID}
You can obtain a valid bookmark ID by choosing the Share option for the bookmark, as described here.
